作者按
本章节主要讲解了基于大模型的自动注释方法,包括CellTypist(发表在Science)和MetaTiME(发表在Nature communication),一个通用,一个泛癌专用。本教程首发于单细胞最好的中文教程,未经授权许可,禁止转载。
全文字数|预计阅读时间: 3000|3min
——Starlitnightly(星夜)
1. 背景
我们在3-3中介绍了基于marker的自动注释方法SCSA,在本章中,我们将介绍基于深度学习模型的自动注释方法CellTypist。该算法发表在Science,算是唯一一篇发表在正刊的单细胞注释算法。与别的自动注释算法不同,CellTypist可以自定义高精度和低精度,也就是说,CellTypist可以直接注释出细胞的亚群,在高精度下。此外,对于不认识的细胞类型,其会选择性注释,不会瞎注释,这与SCSA是相同的。
我们还介绍了一种关于癌症的细胞类型的自动注释算法Metatime,这是基于23种泛癌百万单细胞图谱所构建出来的自动注释模型
import omicverse as ov
print(f'omicverse version: {ov.__version__}')
import scanpy as sc
print(f'scanpy version: {sc.__version__}')
ov.ov_plot_set()
import celltypist
print(f'celltypist version: {celltypist.__version__}')
from celltypist import models
omicverse version: 1.5.1
scanpy version: 1.9.1
celltypist version: 1.6.0
2. 加载数据
我们还是使用前面用到的已经手动注释好的单细胞测序数据
adata = ov.utils.read('data/s4d8_manual_annotation.h5ad')
由于Celltypist要求使用的归一化值为10,000,而omicverse自带预处理的归一化值为500,000。所以我们提取原始counts进行还原,然后重新归一化
adata=adata.raw.to_adata()
ov.utils.retrieve_layers(adata,layers='counts')
print('raw count adata:',adata.X.max())
......The X of adata have been stored in raw
......The layers counts of adata have been retreved
raw count adata: 889.0
adata=ov.pp.preprocess(adata,mode='shiftlog|pearson',n_HVGs=2000,target_sum=1e4)
Begin robust gene identification
After filtration, 20171/20171 genes are kept. Among 20171 genes, 20171 genes are robust.
End of robust gene identification.
Begin size normalization: shiftlog and HVGs selection pearson
normalizing counts per cell The following highly-expressed genes are not considered during normalization factor computation:
[]
finished (0:00:00)
WARNING: adata.X seems to be already log-transformed.
extracting highly variable genes
--> added
'highly_variable', boolean vector (adata.var)
'highly_variable_rank', float vector (adata.var)
'highly_variable_nbatches', int vector (adata.var)
'highly_variable_intersection', boolean vector (adata.var)
'means', float vector (adata.var)
'variances', float vector (adata.var)
'residual_variances', float vector (adata.var)
End of size normalization: shiftlog and HVGs selection pearson
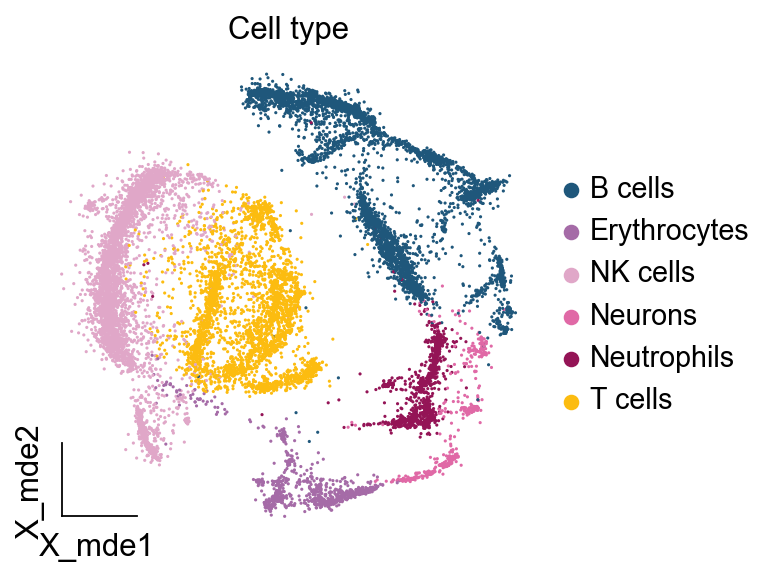
ov.utils.embedding(adata,
basis='X_mde',
color=[ "major_celltype"],
title=['Cell type'],
palette=ov.palette()[11:],
show=False,frameon='small',)
3. CellTypist模型加载
我们首先需要下载已经训练好的模型
# Enabling `force_update = True` will overwrite existing (old) models.
models.download_models(force_update = True)
标签:Downloading,Science,31,测序,注释,model,adata,pkl
From: https://www.cnblogs.com/starlitnightly/p/18258593