转载 医学和生信笔记公众号
医学和生信笔记公众号主要分享4大块内容:
- 生信数据挖掘
- 医学统计分析
- 机器学习
- 临床预测模型
前期主要是以医学统计和临床预测模型为主,关于生信挖掘和机器学习的内容偏少,所以后面会逐渐增加这方面的内容,除了常见的生信分析外,还会涉及一些SCI图表学习等内容。
富集分析作为了解基因功能的常用方法,也是各种生信相关SCI中的常客,基本上是必会的内容。
今天为大家带来常见的富集分析类型,其实主要就是两类:
ORA(Over-Representation Analysis),通过超几何分布检验实现GSEA(gene set enrichment analysis),基因集富集分析
除此之外,还有ssGSEA、GSVA等,我们等下次再介绍。富集分析的可视化,我们也是放到下次再介绍,因为1条推文根本说不完,太多了!
目前做ORA和GSEA,肯定是首选clusterProfiler了,不管是使用的便捷性,还是对各种富集分析的完美支持,还是对各种最新数据库的支持,clusterProfiler都是最好的选择!
关于各种富集分析的底层原理,我们这里就不说了,大家感兴趣的可以自己学习下。
library(clusterProfiler)
##
## clusterProfiler v4.6.2 For help: https://yulab-smu.top/biomedical-knowledge-mining-book/
##
## If you use clusterProfiler in published research, please cite:
## T Wu, E Hu, S Xu, M Chen, P Guo, Z Dai, T Feng, L Zhou, W Tang, L Zhan, X Fu, S Liu, X Bo, and G Yu. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. The Innovation. 2021, 2(3):100141
##
## Attaching package: 'clusterProfiler'
## The following object is masked from 'package:stats':
##
## filterGO富集分析
clusterProfiler为我们提供了3种GO富集分析:
- GO分类
- GO ORA
- GO GSEA
GO分类
基于特定GO分布对基因进行分类,使用起来超级简单,下面我们用示例数据为大家演示。
data(geneList, package="DOSE")
gene <- names(geneList)[abs(geneList) > 2]
# Entrez gene ID
head(gene)
## [1] "4312" "8318" "10874" "55143" "55388" "991"geneList是一个有序的数值型向量,其名字是基因的ID(这里是ENTREZID)。
除了做GSEA分析需要geneList这种格式外,其他都是直接提供基因ID的向量即可:
ggo <- groupGO(gene = gene,
keyType = "ENTREZID",#任意类型都可,不一定非要ENTREZID,这里指定即可
OrgDb = "org.Hs.eg.db",
ont = "CC",# “BP”, “CC”, “MF”
level = 3,
readable = TRUE
)
##
head(ggo)
## ID Description Count GeneRatio geneID
## GO:0000133 GO:0000133 polarisome 0 0/207
## GO:0000408 GO:0000408 EKC/KEOPS complex 0 0/207
## GO:0000417 GO:0000417 HIR complex 0 0/207
## GO:0000444 GO:0000444 MIS12/MIND type complex 0 0/207
## GO:0000808 GO:0000808 origin recognition complex 0 0/207
## GO:0000930 GO:0000930 gamma-tubulin complex 0 0/207GO过代表分析
这个就是大家最常见的GO富集分析了,使用enrichGO函数实现,各种参数也是和上面差不多:
ego <- enrichGO(gene = gene,
universe = names(geneList),#背景基因
OrgDb = "org.Hs.eg.db",
ont = "ALL",#"BP", "MF", "CC"
pAdjustMethod = "BH",
pvalueCutoff = 0.01,
qvalueCutoff = 0.05,
readable = TRUE)
ego
## #
## # over-representation test
## #
## #...@organism Homo sapiens
## #...@ontology GOALL
## #...@keytype ENTREZID
## #...@gene chr [1:207] "4312" "8318" "10874" "55143" "55388" "991" "6280" "2305" ...
## #...pvalues adjusted by 'BH' with cutoff <0.01
## #...153 enriched terms found
## 'data.frame': 153 obs. of 10 variables:
## $ ONTOLOGY : chr "BP" "BP" "BP" "BP" ...
## $ ID : chr "GO:0140014" "GO:0000280" "GO:0000070" "GO:0048285" ...
## $ Description: chr "mitotic nuclear division" "nuclear division" "mitotic sister chromatid segregation" "organelle fission" ...
## $ GeneRatio : chr "32/195" "36/195" "26/195" "37/195" ...
## $ BgRatio : chr "263/11590" "357/11590" "165/11590" "391/11590" ...
## $ pvalue : num 7.10e-19 1.96e-18 2.60e-18 5.24e-18 6.54e-18 ...
## $ p.adjust : num 2.16e-15 2.65e-15 2.65e-15 3.99e-15 3.99e-15 ...
## $ qvalue : num 2.00e-15 2.44e-15 2.44e-15 3.68e-15 3.68e-15 ...
## $ geneID : chr 省略
head(ego)
## ONTOLOGY ID Description GeneRatio
## GO:0140014 BP GO:0140014 mitotic nuclear division 32/195
## GO:0000280 BP GO:0000280 nuclear division 36/195
## GO:0000070 BP GO:0000070 mitotic sister chromatid segregation 26/195
## GO:0048285 BP GO:0048285 organelle fission 37/195
## GO:0000819 BP GO:0000819 sister chromatid segregation 27/195
## GO:0007059 BP GO:0007059 chromosome segregation 30/195
## BgRatio pvalue p.adjust qvalue
## GO:0140014 263/11590 7.101540e-19 2.163839e-15 1.998149e-15
## GO:0000280 357/11590 1.956617e-18 2.645070e-15 2.442530e-15
## GO:0000070 165/11590 2.604269e-18 2.645070e-15 2.442530e-15
## GO:0048285 391/11590 5.235148e-18 3.987643e-15 3.682300e-15
## GO:0000819 188/11590 6.543556e-18 3.987643e-15 3.682300e-15
## GO:0007059 278/11590 2.850765e-16 1.447713e-13 1.336859e-13
## geneID
## GO:0140014 CDCA8/CDC20/KIF23/CENPE/MYBL2/NDC80/NCAPH/DLGAP5/UBE2C/NUSAP1/TPX2/TACC3/NEK2/UBE2S/MAD2L1/KIF18A/CDT1/BIRC5/KIF11/TTK/NCAPG/AURKB/CHEK1/TRIP13/PRC1/KIFC1/KIF18B/AURKA/CCNB1/KIF4A/PTTG1/BMP4
#省略
## Count
## GO:0140014 32
## GO:0000280 36
## GO:0000070 26
## GO:0048285 37
## GO:0000819 27
## GO:0007059 30GO的数据库条目可能有很多是在解释同一个东西,所clusterProfielr提供了simplify能够去除冗余的GO:
simple_ego <- simplify(ego)目前这个函数支持来自enrichGO、gseGO、compareCluster的结果,非常实用!
GO基因集富集分析
GO版本的GSEA富集分析, gene set enrichment analysis,通过gseGO函数实现:
ego3 <- gseGO(geneList = geneList,
OrgDb = "org.Hs.eg.db",
ont = "CC",
minGSSize = 100,
maxGSSize = 500,# 只有基因集数量在[minGSSize, maxGSSize]之间的才会被检验
pvalueCutoff = 0.05,
verbose = FALSE)
head(ego3)
## ID Description setSize
## GO:0000775 GO:0000775 chromosome, centromeric region 188
## GO:0000779 GO:0000779 condensed chromosome, centromeric region 138
## GO:0000776 GO:0000776 kinetochore 130
## GO:0000228 GO:0000228 nuclear chromosome 175
## GO:0098687 GO:0098687 chromosomal region 305
## GO:0000793 GO:0000793 condensed chromosome 210
## enrichmentScore NES pvalue p.adjust qvalue rank
## GO:0000775 0.5970689 2.652931 1e-10 1.62e-09 1.073684e-09 530
## GO:0000779 0.6216009 2.648130 1e-10 1.62e-09 1.073684e-09 798
## GO:0000776 0.6230073 2.609292 1e-10 1.62e-09 1.073684e-09 449
## GO:0000228 0.5875327 2.580497 1e-10 1.62e-09 1.073684e-09 1905
## GO:0098687 0.5419949 2.545830 1e-10 1.62e-09 1.073684e-09 1721
## GO:0000793 0.5507360 2.468350 1e-10 1.62e-09 1.073684e-09 1721
## leading_edge
## GO:0000775 tags=20%, list=4%, signal=19%
## GO:0000779 tags=24%, list=6%, signal=23%
## GO:0000776 tags=21%, list=4%, signal=20%
## GO:0000228 tags=34%, list=15%, signal=29%
## GO:0098687 tags=27%, list=14%, signal=24%
## GO:0000793 tags=28%, list=14%, signal=24%
## 省略前面说过,做GSEA富集分析对数据格式是有要求的:
- 按照递减排序的数值型向量,
- 向量的名字是gene id
KEGG富集分析
KEGG自2012年起不再免费提供FTP下载服务,但是clusterProfiler包支持使用KEGG网站下载最新的在线版本的KEGG数据,KEGG pathway和module都支持哦。
KEGG作为一个常用的数据库是有官方网站的,大家可以自己去熟悉一下:https://www.kegg.jp/
KEGG pathway ORA
KEGG通路ORA分析,KEGG pathway over-representation analysis,通过enrichKEGG函数实现,这些函数的名字都是有规律的,方便你记忆和使用:
kk <- enrichKEGG(gene = gene,
organism = 'hsa',
pvalueCutoff = 0.05)
## Reading KEGG annotation online: "https://rest.kegg.jp/link/hsa/pathway"...
## Reading KEGG annotation online: "https://rest.kegg.jp/list/pathway/hsa"...
head(kk)
## ID Description
## hsa04110 hsa04110 Cell cycle
## hsa04114 hsa04114 Oocyte meiosis
## hsa04218 hsa04218 Cellular senescence
## hsa04061 hsa04061 Viral protein interaction with cytokine and cytokine receptor
## hsa03320 hsa03320 PPAR signaling pathway
## hsa04814 hsa04814 Motor proteins
## GeneRatio BgRatio pvalue p.adjust qvalue
## hsa04110 15/106 157/8465 8.177242e-10 1.717221e-07 1.695702e-07
## hsa04114 10/106 131/8465 5.049610e-06 5.302091e-04 5.235648e-04
## hsa04218 10/106 156/8465 2.366003e-05 1.639157e-03 1.618617e-03
## hsa04061 8/106 100/8465 3.326461e-05 1.639157e-03 1.618617e-03
## hsa03320 7/106 75/8465 3.902756e-05 1.639157e-03 1.618617e-03
## hsa04814 10/106 193/8465 1.433387e-04 5.016856e-03 4.953988e-03
## geneID
## hsa04110 8318/991/9133/10403/890/983/4085/81620/7272/9212/1111/9319/891/4174/9232
## hsa04114 991/9133/983/4085/51806/6790/891/9232/3708/5241
## hsa04218 2305/4605/9133/890/983/51806/1111/891/776/3708
## hsa04061 3627/10563/6373/4283/6362/6355/9547/1524
## hsa03320 4312/9415/9370/5105/2167/3158/5346
## hsa04814 9493/1062/81930/3832/3833/146909/10112/24137/4629/7802
## Count
## hsa04110 15
## hsa04114 10
## hsa04218 10
## hsa04061 8
## hsa03320 7
## hsa04814 10KEGG pathway GSEA
KEGG通路基因集富集分析,KEGG pathway gene set enrichment analysis,通过gseKEGG实现:
kk2 <- gseKEGG(geneList = geneList,
organism = 'hsa',
minGSSize = 120,
pvalueCutoff = 0.05,
verbose = FALSE)
head(kk2)
## ID Description setSize
## hsa04110 hsa04110 Cell cycle 139
## hsa05169 hsa05169 Epstein-Barr virus infection 193
## hsa04613 hsa04613 Neutrophil extracellular trap formation 130
## hsa05166 hsa05166 Human T-cell leukemia virus 1 infection 202
## hsa04510 hsa04510 Focal adhesion 192
## hsa04218 hsa04218 Cellular senescence 141
## enrichmentScore NES pvalue p.adjust qvalue rank
## hsa04110 0.6637551 2.876742 1.000000e-10 7.900000e-09 5.263158e-09 1155
## hsa05169 0.4335010 1.956648 2.185378e-07 8.632244e-06 5.750996e-06 2820
## hsa04613 0.4496569 1.910465 7.157902e-06 1.884914e-04 1.255772e-04 2575
## hsa05166 0.3893613 1.765888 1.482811e-05 2.928551e-04 1.951067e-04 1955
## hsa04510 -0.4233234 -1.741010 3.210562e-05 5.072688e-04 3.379539e-04 2183
## hsa04218 0.4115945 1.788002 8.805195e-05 1.159351e-03 7.723855e-04 1155
## leading_edge
## hsa04110 tags=36%, list=9%, signal=33%
## hsa05169 tags=39%, list=23%, signal=31%
## hsa04613 tags=37%, list=21%, signal=30%
## hsa05166 tags=26%, list=16%, signal=22%
## hsa04510 tags=28%, list=17%, signal=23%
## hsa04218 tags=17%, list=9%, signal=16%
## core_enrichment
## hsa04110 8318/991/9133/10403/890/983/4085/81620/7272/9212/1111/9319/891/4174/9232/4171/993/990/5347/701/9700/898/23594/4998/9134/4175/4173/10926/6502/994/699/4609/5111/26271/1869/1029/8317/4176/2810/3066/1871/1031/9088/995/1019/4172/5885/11200/7027/1875
#省略KEGG module ORA
KEGG模块ORA分析,KEGG module over-representation analysis,通过enrichMKEGG实现:
mkk <- enrichMKEGG(gene = gene,
organism = 'hsa',
pvalueCutoff = 1,
qvalueCutoff = 1)
## Reading KEGG annotation online: "https://rest.kegg.jp/link/hsa/module"...
## Reading KEGG annotation online: "https://rest.kegg.jp/list/module"...
head(mkk)
## ID
## M00912 M00912
## M00095 M00095
## M00053 M00053
## M00938 M00938
## M00003 M00003
## M00049 M00049
## Description
## M00912 NAD biosynthesis, tryptophan => quinolinate => NAD
## M00095 C5 isoprenoid biosynthesis, mevalonate pathway
## M00053 Deoxyribonucleotide biosynthesis, ADP/GDP/CDP/UDP => dATP/dGTP/dCTP/dUTP
## M00938 Pyrimidine deoxyribonucleotide biosynthesis, UDP => dTTP
## M00003 Gluconeogenesis, oxaloacetate => fructose-6P
## M00049 Adenine ribonucleotide biosynthesis, IMP => ADP,ATP
## GeneRatio BgRatio pvalue p.adjust qvalue geneID Count
## M00912 2/9 12/852 0.00620266 0.03721596 0.03264558 23475/3620 2
## M00095 1/9 10/852 0.10126209 0.18449380 0.16183667 3158 1
## M00053 1/9 11/852 0.11086855 0.18449380 0.16183667 6241 1
## M00938 1/9 14/852 0.13914269 0.18449380 0.16183667 6241 1
## M00003 1/9 18/852 0.17559737 0.18449380 0.16183667 5105 1
## M00049 1/9 19/852 0.18449380 0.18449380 0.16183667 26289 1KEGG module GSEA
KEGG模块GSEA分析,KEGG module gene set enrichment analysis
mkk2 <- gseMKEGG(geneList = geneList,
organism = 'hsa',
pvalueCutoff = 1)
## preparing geneSet collections...
## GSEA analysis...
## leading edge analysis...
## done...
head(mkk2)
## ID Description
## M00001 M00001 Glycolysis (Embden-Meyerhof pathway), glucose => pyruvate
## M00938 M00938 Pyrimidine deoxyribonucleotide biosynthesis, UDP => dTTP
## M00002 M00002 Glycolysis, core module involving three-carbon compounds
## M00035 M00035 Methionine degradation
## M00104 M00104 Bile acid biosynthesis, cholesterol => cholate/chenodeoxycholate
## M00009 M00009 Citrate cycle (TCA cycle, Krebs cycle)
## setSize enrichmentScore NES pvalue p.adjust qvalue rank
## M00001 24 0.5739036 1.746110 0.006607415 0.2180447 0.2086552 2886
## M00938 10 0.6648004 1.550423 0.041882396 0.4607064 0.4408673 648
## M00002 11 0.6421781 1.548820 0.039734634 0.4607064 0.4408673 1381
## M00035 11 0.5824213 1.404697 0.090476190 0.6366300 0.6092153 1555
## M00104 10 -0.5876900 -1.370515 0.108247423 0.6366300 0.6092153 961
## M00009 22 0.4504911 1.340658 0.125858124 0.6366300 0.6092153 3514
## leading_edge
## M00001 tags=54%, list=23%, signal=42%
## M00938 tags=40%, list=5%, signal=38%
## M00002 tags=55%, list=11%, signal=49%
## M00035 tags=45%, list=12%, signal=40%
## M00104 tags=50%, list=8%, signal=46%
## M00009 tags=50%, list=28%, signal=36%
## core_enrichment
## M00001 5214/3101/2821/7167/2597/5230/2023/5223/5315/3099/5232/2027/5211
## M00938 6241/7298/4830/1841
## M00002 7167/2597/5230/2023/5223/5315
## M00035 875/1789/191/1788/1786
## M00104 6342/10998/1581/3295/8309
## M00009 3418/50/4190/3419/2271/3421/55753/3417/1431/6389/4191KEGG通路可视化
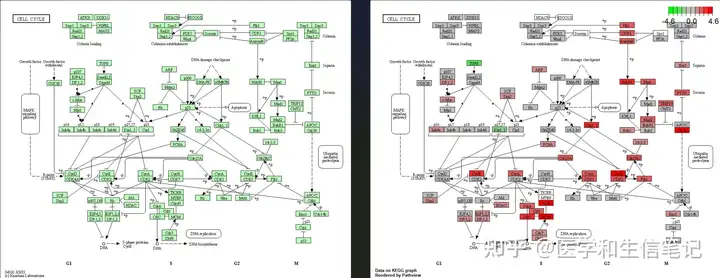
执行以下代码会直接打开网页查看:
browseKEGG(kk, 'hsa04110')或者也可以下载到本地,通过pathview这个包实现:
library("pathview")
hsa04110 <- pathview(gene.data = geneList,
pathway.id = "hsa04110",
species = "hsa",
limit = list(gene=max(abs(geneList)), cpd=1))
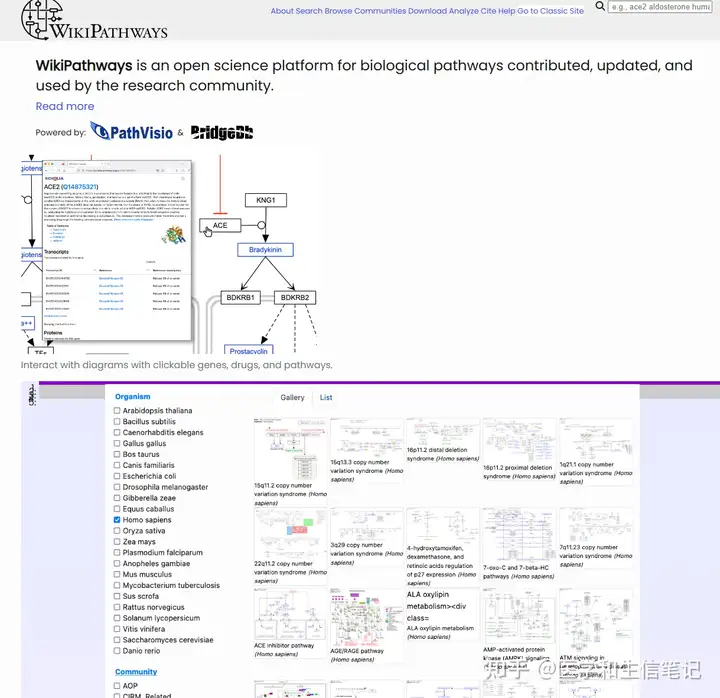
WikiPathways富集分析
WikiPathways(https://www.wikipathways.org/)和KEGG类似,也是一个通路数据库。
富集分析本质上是对基因注释,以了解这些基因的功能,不同的数据库有不同的侧重点,可能今天说的这些注释数据库有几个你没听过,但是不要紧,你可以自己去了解下即可,如果它们都不符合你的要求,你可以自己定义一个注释数据!
WikiPathways ORA
通过enrichWP函数实现,不过需要良好的网络环境,不然就会报错~
enrichWP(gene, organism = "Homo sapiens")
## #
## # over-representation test
## #
## #...@organism Homo sapiens
## #...@ontology WikiPathways
## #...@keytype ENTREZID
## #...@gene chr [1:207] "4312" "8318" "10874" "55143" "55388" "991" "6280" "2305" ...
## #...pvalues adjusted by 'BH' with cutoff <0.05
## #...8 enriched terms found
## 'data.frame': 8 obs. of 9 variables:
## $ ID : chr "WP2446" "WP2361" "WP179" "WP3942" ...
## $ Description: chr "Retinoblastoma gene in cancer" "Gastric cancer network 1" "Cell cycle" "PPAR signaling pathway" ...
## $ GeneRatio : chr "11/113" "6/113" "10/113" "7/113" ...
## $ BgRatio : chr "90/8444" "28/8444" "120/8444" "68/8444" ...
## $ pvalue : num 2.59e-08 1.49e-06 4.18e-06 3.15e-05 3.70e-05 ...
## $ p.adjust : num 7.14e-06 2.06e-04 3.84e-04 2.04e-03 2.04e-03 ...
## $ qvalue : num 6.42e-06 1.85e-04 3.46e-04 1.84e-03 1.84e-03 ...
## $ geneID : chr "8318/9133/7153/6241/890/983/81620/7272/1111/891/24137" "4605/7153/11065/22974/6286/6790" "8318/991/9133/890/983/7272/1111/891/4174/9232" "4312/9415/9370/5105/2167/3158/5346" ...
## $ Count : int 11 6 10 7 4 7 8 10
## #...Citation
## T Wu, E Hu, S Xu, M Chen, P Guo, Z Dai, T Feng, L Zhou, W Tang, L Zhan, X Fu, S Liu, X Bo, and G Yu.
## clusterProfiler 4.0: A universal enrichment tool for interpreting omics data.
## The Innovation. 2021, 2(3):100141WikiPathways gene set enrichment analysis
通过gseWP实现,也是需要良好的网络环境:
gseWP(geneList, organism = "Homo sapiens")
## preparing geneSet collections...
## GSEA analysis...
## leading edge analysis...
## done...
## #
## # Gene Set Enrichment Analysis
## #
## #...@organism Homo sapiens
## #...@setType WikiPathways
## #...@keytype ENTREZID
## #...@geneList Named num [1:12495] 4.57 4.51 4.42 4.14 3.88 ...
## - attr(*, "names")= chr [1:12495] "4312" "8318" "10874" "55143" ...
## #...nPerm
## #...pvalues adjusted by 'BH' with cutoff <0.05
## #...66 enriched terms found
## 'data.frame': 66 obs. of 11 variables:
## $ ID : chr "WP2446" "WP179" "WP466" "WP2361" ...
## $ Description : chr "Retinoblastoma gene in cancer" "Cell cycle" "DNA replication" "Gastric cancer network 1" ...
## $ setSize : int 84 111 42 23 62 201 77 102 41 61 ...
## $ enrichmentScore: num 0.731 0.663 0.792 0.837 0.665 ...
## $ NES : num 2.86 2.75 2.74 2.55 2.47 ...
## $ pvalue : num 1.00e-10 1.00e-10 1.00e-10 1.41e-09 2.66e-09 ...
## $ p.adjust : num 2.12e-08 2.12e-08 2.12e-08 2.24e-07 2.83e-07 ...
## $ qvalue : num 1.77e-08 1.77e-08 1.77e-08 1.87e-07 2.35e-07 ...
## $ rank : num 1333 1234 1002 854 1111 ...
## $ leading_edge : chr "tags=51%, list=11%, signal=46%" "tags=40%, list=10%, signal=36%" "tags=55%, list=8%, signal=51%" "tags=52%, list=7%, signal=49%" ...
## $ core_enrichment: chr "8318/9133/7153/6241/890/983/81620/7272/1111/891/24137/993/898/4998/10733/9134/4175/4173/6502/5984/994/7298/3015"| __truncated__ "8318/991/9133/890/983/7272/1111/891/4174/9232/4171/993/990/5347/9700/898/23594/4998/9134/4175/4173/10926/6502/9"| __truncated__ "8318/55388/81620/4174/4171/990/23594/4998/4175/4173/10926/5984/5111/51053/8317/5427/23649/4176/5982/5557/5558/4172/5424" "4605/7153/11065/22974/6286/6790/1894/56992/4173/1063/9585/8607" ...
## #...Citation
## T Wu, E Hu, S Xu, M Chen, P Guo, Z Dai, T Feng, L Zhou, W Tang, L Zhan, X Fu, S Liu, X Bo, and G Yu.
## clusterProfiler 4.0: A universal enrichment tool for interpreting omics data.
## The Innovation. 2021, 2(3):100141Reactome富集分析
Reactome(https://reactome.org/)也是一个通路数据库~这个网站自带的可视化功能其实很强大,大家可以去试试。
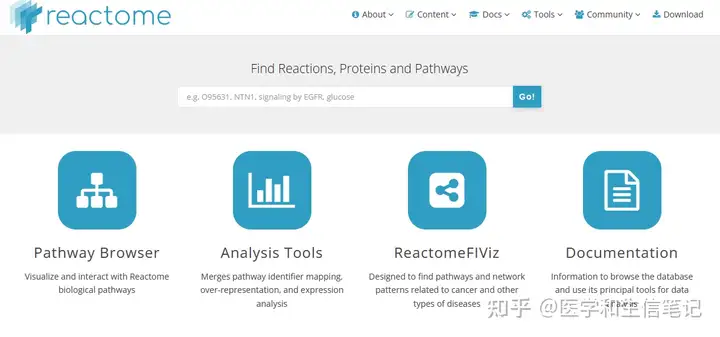
Reactome富集分析是通过ReactomePA包实现的,注意要先加载R包。
Reactome pathway over-representation analysis
Reactome ORA分析,通过函数enrichPathway实现:
library(ReactomePA)
## ReactomePA v1.42.0 For help: https://yulab-smu.top/biomedical-knowledge-mining-book/
##
## If you use ReactomePA in published research, please cite:
## Guangchuang Yu, Qing-Yu He. ReactomePA: an R/Bioconductor package for reactome pathway analysis and visualization. Molecular BioSystems 2016, 12(2):477-479
x <- enrichPathway(gene=gene,
pvalueCutoff = 0.05,
readable=TRUE)
head(x)
## ID Description GeneRatio
## R-HSA-69620 R-HSA-69620 Cell Cycle Checkpoints 22/143
## R-HSA-2500257 R-HSA-2500257 Resolution of Sister Chromatid Cohesion 15/143
## R-HSA-68877 R-HSA-68877 Mitotic Prometaphase 18/143
## R-HSA-69618 R-HSA-69618 Mitotic Spindle Checkpoint 14/143
## R-HSA-68882 R-HSA-68882 Mitotic Anaphase 18/143
## R-HSA-2555396 R-HSA-2555396 Mitotic Metaphase and Anaphase 18/143
## BgRatio pvalue p.adjust qvalue
## R-HSA-69620 293/10899 2.906024e-11 1.330959e-08 1.095112e-08
## R-HSA-2500257 126/10899 8.316509e-11 1.904481e-08 1.567005e-08
## R-HSA-68877 204/10899 1.539628e-10 2.350499e-08 1.933989e-08
## R-HSA-69618 113/10899 2.130715e-10 2.439669e-08 2.007358e-08
## R-HSA-68882 236/10899 1.657466e-09 1.354116e-07 1.114166e-07
## R-HSA-2555396 237/10899 1.773951e-09 1.354116e-07 1.114166e-07
## 省略Reactome pathway gene set enrichment analysis
Reactome GSEA分析,通过函数gsePathway实现:
y <- gsePathway(geneList,
pvalueCutoff = 0.2,
pAdjustMethod = "BH",
verbose = FALSE)
head(y)
## ID Description setSize
## R-HSA-69620 R-HSA-69620 Cell Cycle Checkpoints 237
## R-HSA-453279 R-HSA-453279 Mitotic G1 phase and G1/S transition 142
## R-HSA-69278 R-HSA-69278 Cell Cycle, Mitotic 457
## R-HSA-69306 R-HSA-69306 DNA Replication 137
## R-HSA-69206 R-HSA-69206 G1/S Transition 124
## R-HSA-69481 R-HSA-69481 G2/M Checkpoints 134
## enrichmentScore NES pvalue p.adjust qvalue rank
## R-HSA-69620 0.6678105 3.064037 1e-10 4.105556e-09 3.064327e-09 1905
## R-HSA-453279 0.7016040 3.001137 1e-10 4.105556e-09 3.064327e-09 1155
## R-HSA-69278 0.6077642 2.974757 1e-10 4.105556e-09 3.064327e-09 1769
## R-HSA-69306 0.6968511 2.961013 1e-10 4.105556e-09 3.064327e-09 1769
## R-HSA-69206 0.7046866 2.949856 1e-10 4.105556e-09 3.064327e-09 1155
## R-HSA-69481 0.6876905 2.925269 1e-10 4.105556e-09 3.064327e-09 1905
## leading_edge
## R-HSA-69620 tags=43%, list=15%, signal=37%
## R-HSA-453279 tags=41%, list=9%, signal=37%
## R-HSA-69278 tags=36%, list=14%, signal=32%
## R-HSA-69306 tags=49%, list=14%, signal=42%
## R-HSA-69206 tags=42%, list=9%, signal=38%
## R-HSA-69481 tags=49%, list=15%, signal=42% core_enrichment
## R-HSA-69620 8318/55143/55388/991/1062/9133/10403/11065/220134/79019/55839/27338/890/983/54821/4085/81930/332/9212/1111/891/4174/4171/11004/993/990/5347/701/51512/9156/11130/79682/57405/2491/898/23594/4998/9134/4175/4173/10926/5984/1058/699/1063/85236/5688/5709/55055/641/1029/5698/1763/8970/5693/8317/4176/5713/79980/5982/9735/5721/2810/5691/9088/995/5685/7468/4172/7336/5690/5684/83990/5686/5695/11200/10213/8345/7534/80010/23198/5983/7979/54908/6396/4683/63967/3018/5699/5714/5702/3014/5905/3619/5708/55166/5692/10393/8290/5704/580/6119
## R-HSA-453279 省略 可视化
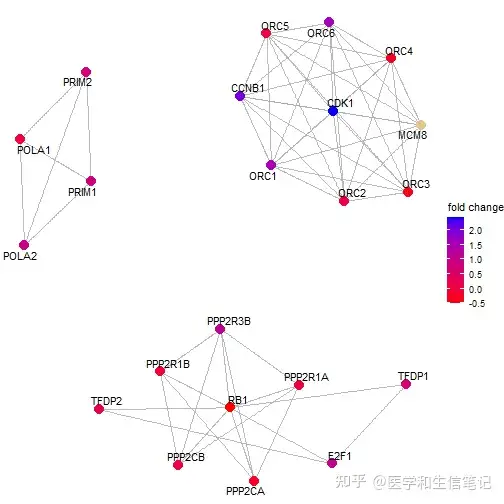
这里简单说下Reactome富集分析的可视化,因为viewPathway函数是它专用的:
viewPathway("E2F mediated regulation of DNA replication",
readable = TRUE,
foldChange = geneList)
## Loading required package: graphite
疾病富集分析
大家比较常用的是对基因做富集分析,但是疾病也是可以做富集分析的,有基因本体论(GO),也有疾病本体论(DO)
DOSE包是专门用来做疾病富集分析的。DOSE提供了五种方法来测量DO-term和基因产物之间的语义相似性,也是通过ORA和GSEA将疾病与基因相关联。
library(DOSE)
## DOSE v3.24.2 For help: https://yulab-smu.top/biomedical-knowledge-mining-book/
##
## If you use DOSE in published research, please cite:
## Guangchuang Yu, Li-Gen Wang, Guang-Rong Yan, Qing-Yu He. DOSE: an R/Bioconductor package for Disease Ontology Semantic and Enrichment analysis. Bioinformatics 2015, 31(4):608-609Disease over-representation analysis
DOSE支持疾病本体论(DO)、癌症基因网络和疾病基因网络(DisGeNET)的富集分析。
- Over-representation analysis for disease ontology
疾病本体论(DO)的ORA分析,和GO的ORA分析一模一样:
x <- enrichDO(gene = gene,
ont = "DO",
pvalueCutoff = 0.05,
pAdjustMethod = "BH",
universe = names(geneList),
minGSSize = 10,
maxGSSize = 500,
qvalueCutoff = 0.05,
readable = FALSE)
head(x)
## ID Description GeneRatio BgRatio
## DOID:1107 DOID:1107 esophageal carcinoma 15/157 195/7744
## DOID:5041 DOID:5041 esophageal cancer 15/157 229/7744
## DOID:3459 DOID:3459 breast carcinoma 22/157 461/7744
## DOID:3748 DOID:3748 esophagus squamous cell carcinoma 10/157 131/7744
## DOID:8719 DOID:8719 in situ carcinoma 10/157 132/7744
## DOID:0060071 DOID:0060071 pre-malignant neoplasm 11/157 159/7744
## pvalue p.adjust qvalue
## DOID:1107 8.724913e-06 0.006343012 0.005675786
## DOID:5041 5.832237e-05 0.021200181 0.018970118
## DOID:3459 1.412419e-04 0.033578155 0.030046043
## DOID:3748 3.100853e-04 0.033578155 0.030046043
## DOID:8719 3.297454e-04 0.033578155 0.030046043
## DOID:0060071 3.664508e-04 0.033578155 0.030046043
## geneID
## DOID:1107 4312/6280/3868/6279/8140/890/5918/332/2146/4321/891/2066/3169/8614/11122
## DOID:5041 4312/6280/3868/6279/8140/890/5918/332/2146/4321/891/2066/3169/8614/11122
## DOID:3459 4312/6280/6279/7153/6241/4751/890/4085/332/6286/2146/6790/891/9232/771/4250/2066/3169/10647/5304/5241/10551
## DOID:3748 6280/3868/6279/8140/5918/2146/891/2066/3169/11122
## DOID:8719 6280/7153/6278/10232/332/9212/6790/891/2066/5241
## DOID:0060071 6280/7153/6278/10232/332/9212/4321/6790/891/2066/5241
## Count
## DOID:1107 15
## DOID:5041 15
## DOID:3459 22
## DOID:3748 10
## DOID:8719 10
## DOID:0060071 11注意:ont参数可以是"DO"或"DOLite",DOLite是一个简化后的DO版本,合并了一些冗余的DO term,但是数据不更新了,建议使用ont="DO"
- Over-representation analysis for the network of cancer gene
癌症基因网络的ORA分析:
gene2 <- names(geneList)[abs(geneList) < 3]
head(gene2)
## [1] "8685" "597" "7153" "23397" "6278" "79733"
ncg <- enrichNCG(gene2)
head(ncg)
## ID
## pan-cancer_paediatric pan-cancer_paediatric
## triple_negative_breast_cancer triple_negative_breast_cancer
## bladder_cancer bladder_cancer
## pancreatic_cancer_(all_histologies) pancreatic_cancer_(all_histologies)
## soft_tissue_sarcoma soft_tissue_sarcoma
## paediatric_high-grade_glioma paediatric_high-grade_glioma
## Description
## pan-cancer_paediatric pan-cancer_paediatric
## triple_negative_breast_cancer triple_negative_breast_cancer
## bladder_cancer bladder_cancer
## pancreatic_cancer_(all_histologies) pancreatic_cancer_(all_histologies)
## soft_tissue_sarcoma soft_tissue_sarcoma
## paediatric_high-grade_glioma paediatric_high-grade_glioma
## GeneRatio BgRatio pvalue
## pan-cancer_paediatric 162/2281 183/3177 1.833773e-08
## triple_negative_breast_cancer 71/2281 75/3177 4.290660e-07
## bladder_cancer 97/2281 112/3177 1.253690e-04
## pancreatic_cancer_(all_histologies) 40/2281 42/3177 1.262162e-04
## soft_tissue_sarcoma 26/2281 26/3177 1.742793e-04
## paediatric_high-grade_glioma 25/2281 25/3177 2.434966e-04
## p.adjust qvalue
## pan-cancer_paediatric 1.613721e-06 7.721152e-07
## triple_negative_breast_cancer 1.887890e-05 9.032967e-06
## bladder_cancer 2.776757e-03 1.328592e-03
## pancreatic_cancer_(all_histologies) 2.776757e-03 1.328592e-03
## soft_tissue_sarcoma 3.067315e-03 1.467615e-03
## paediatric_high-grade_glioma 3.073768e-03 1.470702e-03
## geneID
## pan-cancer_paediatric 2146/55353/4609/1029/3575/22806/3418/3066/2120/30012/867/7468/7545/3195/865/64109/4613/613/11177/7490/238/10736/10054/5771/4893/140885/1785/9760/3417/6597/6476/9126/4869/10320/7307/80204/1050/10992/8028/2312/6608/896/894/2196/4849/7023/5093/5079/5293/5727/55181/171017/51322/5781/3718/55294/60/673/8085/5897/4851/1665/51176/1108/7764/10664/6098/2332/2201/6495/3845/7015/1441/2782/64919/4298/23512/8239/29102/6929/8021/6134/6598/4209/5290/22941/8726/207/3717/2033/10716/4928/6932/694/5156/10019/6886/9968/7080/2623/7874/1654/4149/3020/23219/55252/55729/10735/5728/4853/23451/51341/387/3206/6146/79718/2624/63035/3815/171023/23269/25/23592/5896/7403/2260/54880/3716/9203/57178/6777/5789/4297/29072/90/546/120/25836/8289/4345/9611/5925/4763/1997/1499/7157/3399/5295/1387/4602/51564/1027/4005/2322/2078/678/6403/55709/1277/7494/64061/2625
## triple_negative_breast_cancer 省略
## Count
## pan-cancer_paediatric 162
## triple_negative_breast_cancer 71
## bladder_cancer 97
## pancreatic_cancer_(all_histologies) 40
## soft_tissue_sarcoma 26
## paediatric_high-grade_glioma 25- Over-representation analysis for the disease gene network
疾病基因网络的ORA分析,DisGeNET(https://www.disgenet.org/)是一个综合的、全面的基因-疾病数据库,包含基因-疾病关联和snp-基因-疾病关联。
我做网络药理学时经常使用的一个数据库:
疾病-基因关联的富集分析由enrichDGN函数实现,snp-基因-疾病关联的分析由enrichDGNv函数实现。
dgn <- enrichDGN(gene)
head(dgn)
## ID Description GeneRatio BgRatio
## C0007124 C0007124 Noninfiltrating Intraductal Carcinoma 24/204 486/21671
## C3642347 C3642347 Basal-Like Breast Carcinoma 16/204 245/21671
## C1609538 C1609538 Latent Tuberculosis 14/204 183/21671
## C1827293 C1827293 Carcinoma of urinary bladder, invasive 16/204 287/21671
## C0278601 C0278601 Inflammatory Breast Carcinoma 13/204 183/21671
## C4733092 C4733092 estrogen receptor-negative breast cancer 17/204 356/21671
## pvalue p.adjust qvalue
## C0007124 3.806436e-11 1.568251e-07 1.271350e-07
## C3642347 1.555102e-09 3.038993e-06 2.463650e-06
## C1609538 2.212859e-09 3.038993e-06 2.463650e-06
## C1827293 1.491211e-08 1.535947e-05 1.245161e-05
## C0278601 2.050738e-08 1.689808e-05 1.369893e-05
## C4733092 5.070020e-08 3.481414e-05 2.822311e-05
## geneID
## C0007124 4312/6280/6279/7153/6278/9582/55872/3669/4751/5080/2146/1111/3887/6790/9370/125/8839/2066/3169/9547/10647/2625/7021/5241
## C3642347 2305/1062/4605/9833/7368/11065/10232/55765/5163/2146/2568/3620/6790/3169/2625/5241
## C1609538 8685/3627/3669/6373/820/3002/50852/3902/3620/6355/10578/9370/3117/23158
## C1827293 2305/6279/55165/220134/2146/9212/4321/6790/891/8544/9122/23158/652/2625/5241/10551
## C0278601 4312/6278/11065/9582/51203/5918/2146/4321/3887/2066/9547/10647/5241
## C4733092 2305/6278/79733/6241/81930/81620/2146/3620/80129/4137/8839/3169/1408/5304/2625/5241/10551
## Count
## C0007124 24
## C3642347 16
## C1609538 14
## C1827293 16
## C0278601 13
## C4733092 17
snp <- c("rs1401296", "rs9315050", "rs5498", "rs1524668", "rs147377392",
"rs841", "rs909253", "rs7193343", "rs3918232", "rs3760396",
"rs2231137", "rs10947803", "rs17222919", "rs386602276", "rs11053646",
"rs1805192", "rs139564723", "rs2230806", "rs20417", "rs966221")
dgnv <- enrichDGNv(snp)
head(dgnv)
## ID Description GeneRatio BgRatio pvalue
## C0010054 C0010054 Coronary Arteriosclerosis 6/17 440/194515 1.568917e-12
## C0151744 C0151744 Myocardial Ischemia 4/17 103/194515 1.754840e-10
## C0031099 C0031099 Periodontitis 4/17 116/194515 2.839985e-10
## C0007785 C0007785 Cerebral Infarction 4/17 123/194515 3.599531e-10
## C0003850 C0003850 Arteriosclerosis 4/17 267/194515 8.145389e-09
## C0004153 C0004153 Atherosclerosis 4/17 281/194515 9.996713e-09
## p.adjust qvalue
## C0010054 2.761295e-10 NA
## C0151744 1.544259e-08 NA
## C0031099 1.583793e-08 NA
## C0007785 1.583793e-08 NA
## C0003850 2.867177e-07 NA
## C0004153 2.932369e-07 NA
## geneID Count
## C0010054 rs5498/rs147377392/rs11053646/rs1805192/rs2230806/rs20417 6
## C0151744 rs5498/rs147377392/rs11053646/rs1805192 4
## C0031099 rs5498/rs909253/rs1805192/rs20417 4
## C0007785 rs147377392/rs11053646/rs1805192/rs2230806 4
## C0003850 rs5498/rs1805192/rs2230806/rs20417 4
## C0004153 rs5498/rs1805192/rs2230806/rs20417 4Disease gene set enrichment analysis
DO GSEA分析:
y <- gseDO(geneList,
minGSSize = 120,
pvalueCutoff = 0.2,
pAdjustMethod = "BH",
verbose = FALSE)
head(y, 3)
## ID Description setSize
## DOID:399 DOID:399 tuberculosis 181
## DOID:612 DOID:612 primary immunodeficiency disease 305
## DOID:0050338 DOID:0050338 primary bacterial infectious disease 264
## enrichmentScore NES pvalue p.adjust qvalue
## DOID:399 0.5030583 2.252875 1.00000e-10 2.180000e-08 9.789474e-09
## DOID:612 0.4023288 1.898732 4.87282e-09 5.311374e-07 2.385117e-07
## DOID:0050338 0.4150405 1.945371 1.17702e-08 6.414760e-07 2.880602e-07
## rank leading_edge
## DOID:399 2082 tags=40%, list=17%, signal=34%
## DOID:612 2099 tags=33%, list=17%, signal=29%
## DOID:0050338 1864 tags=33%, list=15%, signal=29%
## 省略 癌症基因网络的GSEA分析:
ncg <- gseNCG(geneList,
pvalueCutoff = 0.5,
pAdjustMethod = "BH",
verbose = FALSE)
ncg <- setReadable(ncg, 'org.Hs.eg.db')
head(ncg, 3)
## ID
## pan-gynecological and breast pan-gynecological and breast
## pan-gastric pan-gastric
## esophageal_adenocarcinoma esophageal_adenocarcinoma
## Description setSize
## pan-gynecological and breast pan-gynecological and breast 43
## pan-gastric pan-gastric 49
## esophageal_adenocarcinoma esophageal_adenocarcinoma 92
## enrichmentScore NES pvalue p.adjust
## pan-gynecological and breast -0.5263429 -1.734359 0.004764925 0.1718695
## pan-gastric -0.4993803 -1.683334 0.006365539 0.1718695
## esophageal_adenocarcinoma -0.4235434 -1.550378 0.004980207 0.1718695
## qvalue rank leading_edge
## pan-gynecological and breast 0.1585801 2464 tags=44%, list=20%, signal=36%
## pan-gastric 0.1585801 3280 tags=49%, list=26%, signal=36%
## esophageal_adenocarcinoma 0.1585801 2464 tags=34%, list=20%, signal=27%
## core_enrichment
## pan-gynecological and breast ATM/ZC3H13/NIPBL/SPOP/ARID1A/RASA1/RB1/RNF43/MAP2K4/NF1/CTNNB1/TP53/PIK3R1/CDKN1B/CCND1/ARID5B/MAP3K1/TBX3/GATA3
## pan-gastric BCOR/SOX9/TCF7L2/ATM/CALD1/SEMG2/HTR7/ARID1A/RASA1/RB1/TTBK2/RNF43/CTNNB1/TP53/BCL9/SMAD3/APC/ZFP36L2/TGFBR2/MUC6/MAP3K1/CACNA1C/ATP8B1/CYP4B1
## esophageal_adenocarcinoma NIPBL/ARID1A/RASA1/MSH3/RNF43/CTNNB1/DNAH7/TP53/GATAD1/PIK3R1/STK11/CDKN1B/SPART/APC/SLIT3/LIN7A/SLIT2/TGFBR2/EPHA3/MUC6/ABCB1/NAV3/SYNE1/MAP3K1/SEMA5A/FTO/NUAK1/PCDH9/CHL1/ABCG2/TRIM58疾病基因网络的GSEA分析:
dgn <- gseDGN(geneList,
pvalueCutoff = 0.2,
pAdjustMethod = "BH",
verbose = FALSE)
dgn <- setReadable(dgn, 'org.Hs.eg.db')
head(dgn, 3)
## ID Description setSize enrichmentScore
## C0024266 C0024266 Lymphocytic Choriomeningitis 120 0.5712593
## C4721414 C4721414 Mantle cell lymphoma 368 0.4107437
## C0205682 C0205682 Waist-Hip Ratio 401 -0.4425633
## NES pvalue p.adjust qvalue rank
## C0024266 2.437904 1e-10 2.05275e-07 1.757895e-07 2579
## C4721414 1.951356 1e-10 2.05275e-07 1.757895e-07 1745
## C0205682 -1.946241 1e-10 2.05275e-07 1.757895e-07 2011
## leading_edge
## C0024266 tags=48%, list=21%, signal=38%
## C4721414 tags=26%, list=14%, signal=23%
## C0205682 tags=28%, list=16%, signal=24%
## 省略MeSH富集分析
meshes包支持使用MeSH注释(医学主题词表)对基因列表或整个表达谱进行富集分析(ORA和GSEA)。gendoo/gene2pubmed/RBBH的数据源都支持。
但是进行MeSH富集分析需要通过AnnotationHub下载数据,这一步由于网络问题经常失败...
AnnotationHub在bioconductor 3.16版本(对应R4.2.0版本)之后得到了重用,专门用于存放各种注释数据的R包,通过它可以非常方便的进行查询、下载所有的注释数据!在此之前的版本都是各种注释数据都有自己的专属R包,不管R包多大,我们都可以通过更换镜像实现快速下载!
现在统一之后确实方便大家查找,但是我觉得这样做让大陆用户很为难......基本上都会卡在下载数据这一步(因为这个东西没有镜像可以用了...),即使通过其他方式下载了数据,但是不同的注释数据还有不同的处理方法。
MeSH over-representation analysis
首先自己构建下数据库,AnnotationHub下载数据太慢了...(但是如果网络环境好也是嗖嗖嗖地下载)
# 需要bioc版本在3.14以上
library(AnnotationHub)
library(MeSHDbi)
ah <- AnnotationHub() #第一次运行会问你要不要建立本地缓存,选yes
hsa <- query(ah, c("MeSHDb", "Homo sapiens"))
file_hsa <- hsa[[1]] # 这一步是下载真正的注释,还好不用每次都下载
#file_hsa <- "C:/Users/liyue/AppData/Local/R/cache/R/AnnotationHub/mesh_hs_db_98354"#这个是我自己通过迅雷下载的,可以读取,但是不知道为啥富集分析不能用
db <- MeSHDbi::MeSHDb(file_hsa)来自官方的提示:
MeSH-related packages (MeSH.XXX.eg.db, MeSH.db, MeSH.AOR.db, and MeSH.PCR.db) are deprecated since Bioconductor 3.14. Use AnnotationHub instead. For details, check the vignette of MeSHDbi
以前是可以安装各种注释R包的,换个镜像即可嗖嗖的下载,好日子一去不复返了!
## BioC 2.14 to BioC 3.13 ##
library(MeSH.Hsa.eg.db)
db <- MeSH.Hsa.eg.db接下来就是进行MeSH的ORA分析:
library(meshes)
## meshes v1.24.0
##
## If you use meshes in published research, please cite the most appropriate paper(s):
##
## Guangchuang Yu. Using meshes for MeSH term enrichment and semantic analyses. Bioinformatics 2018, 34(21):3766-3767, doi:10.1093/bioinformatics/bty410
##
## Attaching package: 'meshes'
## The following object is masked from 'package:DOSE':
##
## geneSim
data(geneList, package="DOSE")
de <- names(geneList)[1:100]
x <- enrichMeSH(de, MeSHDb = db, database='gendoo', category = 'C')
## Error in h(simpleError(msg, call)): error in evaluating the argument 'x' in selecting a method for function 'select': object 'db' not found
head(x)
## ID Description GeneRatio BgRatio
## DOID:1107 DOID:1107 esophageal carcinoma 15/157 195/7744
## DOID:5041 DOID:5041 esophageal cancer 15/157 229/7744
## DOID:3459 DOID:3459 breast carcinoma 22/157 461/7744
## DOID:3748 DOID:3748 esophagus squamous cell carcinoma 10/157 131/7744
## DOID:8719 DOID:8719 in situ carcinoma 10/157 132/7744
## DOID:0060071 DOID:0060071 pre-malignant neoplasm 11/157 159/7744
## pvalue p.adjust qvalue
## DOID:1107 8.724913e-06 0.006343012 0.005675786
## DOID:5041 5.832237e-05 0.021200181 0.018970118
## DOID:3459 1.412419e-04 0.033578155 0.030046043
## DOID:3748 3.100853e-04 0.033578155 0.030046043
## DOID:8719 3.297454e-04 0.033578155 0.030046043
## DOID:0060071 3.664508e-04 0.033578155 0.030046043
## geneID
## DOID:1107 4312/6280/3868/6279/8140/890/5918/332/2146/4321/891/2066/3169/8614/11122
## DOID:5041 4312/6280/3868/6279/8140/890/5918/332/2146/4321/891/2066/3169/8614/11122
## DOID:3459 4312/6280/6279/7153/6241/4751/890/4085/332/6286/2146/6790/891/9232/771/4250/2066/3169/10647/5304/5241/10551
## DOID:3748 6280/3868/6279/8140/5918/2146/891/2066/3169/11122
## DOID:8719 6280/7153/6278/10232/332/9212/6790/891/2066/5241
## DOID:0060071 6280/7153/6278/10232/332/9212/4321/6790/891/2066/5241
## Count
## DOID:1107 15
## DOID:5041 15
## DOID:3459 22
## DOID:3748 10
## DOID:8719 10
## DOID:0060071 11MeSH gene set enrichment analysis
然后是GSEA分析:
y <- gseMeSH(geneList, MeSHDb = db, database = 'gene2pubmed', category = "G")
## Error in h(simpleError(msg, call)): error in evaluating the argument 'x' in selecting a method for function 'select': object 'db' not found
head(y)
## ID Description
## DOID:399 DOID:399 tuberculosis
## DOID:612 DOID:612 primary immunodeficiency disease
## DOID:0050338 DOID:0050338 primary bacterial infectious disease
## DOID:104 DOID:104 bacterial infectious disease
## DOID:526 DOID:526 human immunodeficiency virus infectious disease
## DOID:2237 DOID:2237 hepatitis
## setSize enrichmentScore NES pvalue p.adjust
## DOID:399 181 0.5030583 2.252875 1.000000e-10 2.180000e-08
## DOID:612 305 0.4023288 1.898732 4.872820e-09 5.311374e-07
## DOID:0050338 264 0.4150405 1.945371 1.177020e-08 6.414760e-07
## DOID:104 321 0.3899673 1.856122 9.801772e-09 6.414760e-07
## DOID:526 201 0.4147782 1.874250 4.756033e-07 2.073630e-05
## DOID:2237 406 0.3328749 1.613797 1.567985e-06 5.697013e-05
## qvalue rank leading_edge
## DOID:399 9.789474e-09 2082 tags=40%, list=17%, signal=34%
## DOID:612 2.385117e-07 2099 tags=33%, list=17%, signal=29%
## DOID:0050338 2.880602e-07 1864 tags=33%, list=15%, signal=29%
## DOID:104 2.880602e-07 2330 tags=36%, list=19%, signal=30%
## DOID:526 9.311812e-06 2099 tags=36%, list=17%, signal=31%
## DOID:2237 2.558292e-05 2082 tags=27%, list=17%, signal=23%
## 省略自定义富集分析
有些用户需要自定义自己的注释数据库或者没有支持的物种,也可以自己DIY。
clusterProfiler提供了用于超几何检验的enricher()函数和用于基因集富集分析的GSEA()函数,可以让用户根据自己的注释信息进行富集分析。
需要两个额外的参数TERM2GENE和TERM2NAME。TERM2GENE是一个数据框,第一列是term-ID,第二列是相应的映射基因;TERM2NAME也是一个数据框,第一列是term-ID,第二列是相应的术语名称。其中TERM2NAME是可选的。
MiSigdb
下面是一个用MiSigdb数据库进行富集分析的例子。MiSigdb可以非常方便的进行查询和下载MiSigdb数据库的基因集。
查看支持的物种:
library(msigdbr)
msigdbr_show_species()
## [1] "Anolis carolinensis" "Bos taurus"
## [3] "Caenorhabditis elegans" "Canis lupus familiaris"
## [5] "Danio rerio" "Drosophila melanogaster"
## [7] "Equus caballus" "Felis catus"
## [9] "Gallus gallus" "Homo sapiens"
## [11] "Macaca mulatta" "Monodelphis domestica"
## [13] "Mus musculus" "Ornithorhynchus anatinus"
## [15] "Pan troglodytes" "Rattus norvegicus"
## [17] "Saccharomyces cerevisiae" "Schizosaccharomyces pombe 972h-"
## [19] "Sus scrofa" "Xenopus tropicalis"选择我们想要的数据下载:
m_df <- msigdbr(species = "Homo sapiens")#下载数据
head(m_df, 2) %>% as.data.frame
## gs_cat gs_subcat gs_name gene_symbol entrez_gene ensembl_gene
## 1 C3 MIR:MIR_Legacy AAACCAC_MIR140 ABCC4 10257 ENSG00000125257
## 2 C3 MIR:MIR_Legacy AAACCAC_MIR140 ABRAXAS2 23172 ENSG00000165660
## human_gene_symbol human_entrez_gene human_ensembl_gene gs_id gs_pmid
## 1 ABCC4 10257 ENSG00000125257 M12609
## 2 ABRAXAS2 23172 ENSG00000165660 M12609
## gs_geoid gs_exact_source gs_url
## 1
## 2
## gs_description
## 1 Genes having at least one occurence of the motif AAACCAC in their 3' untranslated region. The motif represents putative target (that is, seed match) of human mature miRNA hsa-miR-140 (v7.1 miRBase).
## 2 Genes having at least one occurence of the motif AAACCAC in their 3' untranslated region. The motif represents putative target (that is, seed match) of human mature miRNA hsa-miR-140 (v7.1 miRBase).我们使用人类的C6注释集:
m_t2g <- msigdbr(species = "Homo sapiens", category = "C6") %>%
dplyr::select(gs_name, entrez_gene)
head(m_t2g)
## # A tibble: 6 × 2
## gs_name entrez_gene
## <chr> <int>
## 1 AKT_UP.V1_DN 57007
## 2 AKT_UP.V1_DN 22859
## 3 AKT_UP.V1_DN 22859
## 4 AKT_UP.V1_DN 137872
## 5 AKT_UP.V1_DN 249
## 6 AKT_UP.V1_DN 271有了这个数据之后,我们就可以根据这个自定义的注释信息做富集分析了:
首先是ORA分析:
em <- enricher(gene, TERM2GENE=m_t2g)
head(em)
## ID Description GeneRatio
## RPS14_DN.V1_DN RPS14_DN.V1_DN RPS14_DN.V1_DN 22/183
## GCNP_SHH_UP_LATE.V1_UP GCNP_SHH_UP_LATE.V1_UP GCNP_SHH_UP_LATE.V1_UP 16/183
## PRC2_EZH2_UP.V1_DN PRC2_EZH2_UP.V1_DN PRC2_EZH2_UP.V1_DN 15/183
## VEGF_A_UP.V1_DN VEGF_A_UP.V1_DN VEGF_A_UP.V1_DN 15/183
## RB_P107_DN.V1_UP RB_P107_DN.V1_UP RB_P107_DN.V1_UP 10/183
## E2F1_UP.V1_UP E2F1_UP.V1_UP E2F1_UP.V1_UP 12/183
## BgRatio pvalue p.adjust qvalue
## RPS14_DN.V1_DN 186/10922 4.657342e-13 7.870908e-11 6.422230e-11
## GCNP_SHH_UP_LATE.V1_UP 181/10922 5.765032e-08 4.871452e-06 3.974838e-06
## PRC2_EZH2_UP.V1_DN 192/10922 7.574546e-07 3.419660e-05 2.790255e-05
## VEGF_A_UP.V1_DN 193/10922 8.093869e-07 3.419660e-05 2.790255e-05
## RB_P107_DN.V1_UP 130/10922 6.394609e-05 2.013355e-03 1.642787e-03
## E2F1_UP.V1_UP 188/10922 7.477287e-05 2.013355e-03 1.642787e-03
## geneID
## RPS14_DN.V1_DN 10874/55388/991/9493/1062/4605/9133/23397/79733/9787/55872/83461/54821/51659/9319/9055/10112/4174/5105/2532/7021/79901
## GCNP_SHH_UP_LATE.V1_UP 55388/7153/79733/6241/9787/51203/983/9212/1111/9319/9055/3833/6790/4174/3169/1580
## PRC2_EZH2_UP.V1_DN 8318/55388/4605/23397/9787/55355/10460/6362/81620/2146/7272/9212/11182/3887/24137
## VEGF_A_UP.V1_DN 8318/9493/1062/9133/10403/6241/9787/4085/332/3832/7272/891/23362/2167/10234
## RB_P107_DN.V1_UP 8318/23397/79733/6241/4085/8208/9055/24137/4174/1307
## E2F1_UP.V1_UP 55388/7153/23397/79733/9787/2146/2842/9212/8208/1111/9055/3833
## Count
## RPS14_DN.V1_DN 22
## GCNP_SHH_UP_LATE.V1_UP 16
## PRC2_EZH2_UP.V1_DN 15
## VEGF_A_UP.V1_DN 15
## RB_P107_DN.V1_UP 10
## E2F1_UP.V1_UP 12再试一下C3基因集:
C3_t2g <- msigdbr(species = "Homo sapiens", category = "C3") %>%
dplyr::select(gs_name, entrez_gene)
head(C3_t2g)
## # A tibble: 6 × 2
## gs_name entrez_gene
## <chr> <int>
## 1 AAACCAC_MIR140 10257
## 2 AAACCAC_MIR140 23172
## 3 AAACCAC_MIR140 81
## 4 AAACCAC_MIR140 81
## 5 AAACCAC_MIR140 90
## 6 AAACCAC_MIR140 8754然后是GSEA分析:
em2 <- GSEA(geneList, TERM2GENE = C3_t2g)
## preparing geneSet collections...
## GSEA analysis...
## leading edge analysis...
## done...
head(em2)
## ID Description setSize
## HSD17B8_TARGET_GENES HSD17B8_TARGET_GENES HSD17B8_TARGET_GENES 401
## E2F1_Q3 E2F1_Q3 E2F1_Q3 187
## E2F1_Q6 E2F1_Q6 E2F1_Q6 185
## SGCGSSAAA_E2F1DP2_01 SGCGSSAAA_E2F1DP2_01 SGCGSSAAA_E2F1DP2_01 129
## E2F_Q4 E2F_Q4 E2F_Q4 193
## E2F1DP1_01 E2F1DP1_01 E2F1DP1_01 184
## enrichmentScore NES pvalue p.adjust
## HSD17B8_TARGET_GENES 0.6368099 3.107103 1.000000e-10 1.655000e-07
## E2F1_Q3 0.4983913 2.220028 1.000000e-10 1.655000e-07
## E2F1_Q6 0.4906657 2.184070 2.011165e-10 2.218985e-07
## SGCGSSAAA_E2F1DP2_01 0.5408182 2.289484 1.032436e-09 8.543410e-07
## E2F_Q4 0.4685394 2.099303 2.297950e-09 9.507767e-07
## E2F1DP1_01 0.4675989 2.083101 2.123251e-09 9.507767e-07
## qvalue rank leading_edge
## HSD17B8_TARGET_GENES 1.310526e-07 1042 tags=36%, list=8%, signal=34%
## E2F1_Q3 1.310526e-07 2045 tags=36%, list=16%, signal=30%
## E2F1_Q6 1.757123e-07 1819 tags=34%, list=15%, signal=30%
## SGCGSSAAA_E2F1DP2_01 6.765174e-07 1797 tags=38%, list=14%, signal=33%
## E2F_Q4 7.528809e-07 1663 tags=29%, list=13%, signal=26%
## E2F1DP1_01 7.528809e-07 1797 tags=33%, list=14%, signal=28%
#省略cellmarker
除了msigdb数据库,还有一个cellmarker数据库,做单细胞的一定见过,我们先下载它的注释数据:
cell_marker_data <- vroom::vroom('http://xteam.xbio.top/CellMarker/download/Human_cell_markers.txt')
## Rows: 2868 Columns: 15
## ── Column specification ─────────────────────────────
## Delimiter: "\t"
## chr (15): speciesType, tissueType, UberonOntologyID, cancerType, cellType, c...
##
## ℹ Use `spec()` to retrieve the full column specification for this data.
## ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
## instead of `cellName`, users can use other features (e.g. `cancerType`)
cells <- cell_marker_data %>%
dplyr::select(cellName, geneID) %>%
dplyr::mutate(geneID = strsplit(geneID, ', ')) %>%
tidyr::unnest()有了数据就可以使用通用的enricher函数进行ORA富集分析了:
x <- enricher(gene, TERM2GENE = cells)
head(x)
## ID Description GeneRatio
## Neural progenitor cell Neural progenitor cell Neural progenitor cell 33/153
## MKI67+ progenitor cell MKI67+ progenitor cell MKI67+ progenitor cell 20/153
## DCLK1+ progenitor cell DCLK1+ progenitor cell DCLK1+ progenitor cell 7/153
## BgRatio pvalue p.adjust qvalue
## Neural progenitor cell 176/11582 3.395466e-29 3.259648e-27 2.716373e-27
## MKI67+ progenitor cell 111/11582 1.426062e-17 6.845100e-16 5.704250e-16
## DCLK1+ progenitor cell 99/11582 3.298755e-04 1.055601e-02 8.796679e-03
## geneID
## Neural progenitor cell 991/9493/9833/9133/10403/7153/259266/6241/9787/11065/55355/55872/51203/83461/22974/10460/79019/55839/890/983/4085/5080/332/3832/9212/51659/9055/891/4174/9232/1602/2018/4137
## MKI67+ progenitor cell 991/9133/10403/7153/259266/6241/9787/11065/51203/22974/10460/890/983/332/9212/9055/891/9232/2167/51705
## DCLK1+ progenitor cell 1307/730/54829/10234/79901/57758/4969
## Count
## Neural progenitor cell 33
## MKI67+ progenitor cell 20
## DCLK1+ progenitor cell 7使用GSEA进行基因集富集分析:
y <- GSEA(geneList, TERM2GENE = cells)
## preparing geneSet collections...
## GSEA analysis...
## leading edge analysis...
## done...
head(y)
## ID
## Neural progenitor cell Neural progenitor cell
## FOXN4+ cell FOXN4+ cell
## Paneth cell Paneth cell
## Leydig precursor cell Leydig precursor cell
## FGFR1HighNME5- epithelial cell FGFR1HighNME5- epithelial cell
## Ciliated epithelial cell Ciliated epithelial cell
## Description setSize
## Neural progenitor cell Neural progenitor cell 149
## FOXN4+ cell FOXN4+ cell 63
## Paneth cell Paneth cell 261
## Leydig precursor cell Leydig precursor cell 184
## FGFR1HighNME5- epithelial cell FGFR1HighNME5- epithelial cell 131
## Ciliated epithelial cell Ciliated epithelial cell 217
## enrichmentScore NES pvalue
## Neural progenitor cell 0.7160571 3.085727 1.000000e-10
## FOXN4+ cell 0.7136090 2.685258 1.000000e-10
## Paneth cell 0.5381280 2.489150 1.000000e-10
## Leydig precursor cell -0.5710583 -2.308894 1.000000e-10
## FGFR1HighNME5- epithelial cell -0.5739472 -2.229562 1.476993e-10
## Ciliated epithelial cell -0.5012031 -2.064339 3.336445e-10
## p.adjust qvalue rank
## Neural progenitor cell 4.125000e-09 2.315789e-09 684
## FOXN4+ cell 4.125000e-09 2.315789e-09 1419
## Paneth cell 4.125000e-09 2.315789e-09 2187
## Leydig precursor cell 4.125000e-09 2.315789e-09 2609
## FGFR1HighNME5- epithelial cell 4.874076e-09 2.736323e-09 1404
## Ciliated epithelial cell 9.175223e-09 5.151002e-09 1440
## leading_edge
## Neural progenitor cell tags=47%, list=5%, signal=45%
## FOXN4+ cell tags=51%, list=11%, signal=45%
## Paneth cell tags=45%, list=18%, signal=38%
## Leydig precursor cell tags=45%, list=21%, signal=36%
## FGFR1HighNME5- epithelial cell tags=39%, list=11%, signal=35%
## Ciliated epithelial cell tags=29%, list=12%, signal=27%
#省略DO, GO, Reactome的分析是有readable参数的,其实是根据OrgDb进行转换的,这和setReadable函数的原理一样,所以用到OrgDb的都有这个参数。
多个基因集富集结果比较
同时对多个基因集进行富集分析,这个功能在单细胞里面简直是绝配!
data(gcSample)
str(gcSample)
## List of 8
## $ X1: chr [1:216] "4597" "7111" "5266" "2175" ...
## $ X2: chr [1:805] "23450" "5160" "7126" "26118" ...
## $ X3: chr [1:392] "894" "7057" "22906" "3339" ...
## $ X4: chr [1:838] "5573" "7453" "5245" "23450" ...
## $ X5: chr [1:929] "5982" "7318" "6352" "2101" ...
## $ X6: chr [1:585] "5337" "9295" "4035" "811" ...
## $ X7: chr [1:582] "2621" "2665" "5690" "3608" ...
## $ X8: chr [1:237] "2665" "4735" "1327" "3192" ...同时对这8个基因集分别进行富集分析:
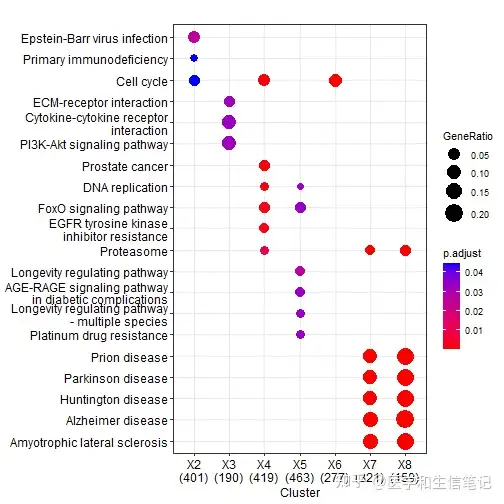
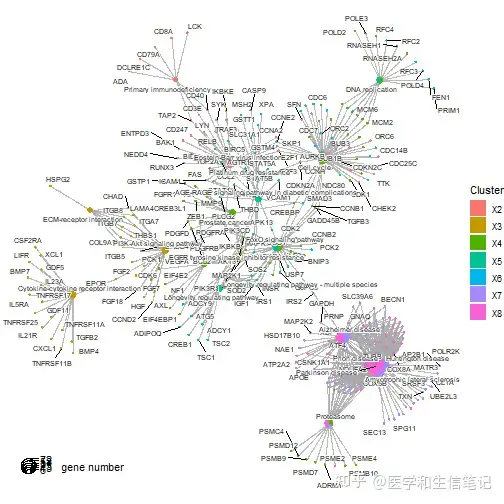
ck <- compareCluster(geneCluster = gcSample, fun = enrichKEGG)
ck <- setReadable(ck, OrgDb = org.Hs.eg.db, keyType="ENTREZID")
head(ck)
## Cluster ID Description GeneRatio BgRatio
## 1 X2 hsa05169 Epstein-Barr virus infection 23/401 202/8465
## 2 X2 hsa05340 Primary immunodeficiency 8/401 38/8465
## 3 X2 hsa04110 Cell cycle 18/401 157/8465
## 4 X3 hsa04512 ECM-receptor interaction 9/190 89/8465
## 5 X3 hsa04060 Cytokine-cytokine receptor interaction 17/190 297/8465
## 6 X3 hsa04151 PI3K-Akt signaling pathway 19/190 354/8465
## pvalue p.adjust qvalue
## 1 7.981195e-05 0.02466189 0.02310346
## 2 3.300857e-04 0.04463551 0.04181490
## 3 4.333545e-04 0.04463551 0.04181490
## 4 1.617209e-04 0.03272190 0.03037690
## 5 3.565676e-04 0.03272190 0.03037690
## 6 3.622350e-04 0.03272190 0.03037690
## geneID
## 1 LYN/ICAM1/TNFAIP3/E2F1/CCNA2/E2F3/BAK1/RUNX3/BID/IKBKE/TAP2/FAS/CCNE2/RELB/CD3E/ENTPD3/SYK/TRAF3/IKBKB/CD247/NEDD4/CD40/STAT1
## 2 ADA/TAP2/LCK/CD79A/CD3E/CD8A/CD40/DCLRE1C
## 3 CDC20/E2F1/CCNA2/E2F3/BUB1B/CDC6/DBF4/PKMYT1/CDC7/ESPL1/CCNE2/CDKN2A/SFN/BUB1/CHEK2/ORC6/CDC14B/MCM4
## 4 THBS1/HSPG2/COL9A3/ITGB7/CHAD/ITGA7/LAMA4/ITGB8/ITGB5
## 5 CXCL1/TNFRSF11B/LIFR/XCL1/GDF5/TNFRSF17/TNFRSF11A/IL5RA/EPOR/CSF2RA/TNFRSF25/BMP7/BMP4/GDF11/IL21R/IL23A/TGFB2
## 6 CCND2/THBS1/STK11/FGF2/COL9A3/ITGB7/FGF7/ERBB4/CHAD/FGF18/CDK6/PCK1/ITGA7/HGF/EPOR/LAMA4/IKBKB/ITGB8/ITGB5
## Count
## 1 23
## 2 8
## 3 18
## 4 9
## 5 17
## 6 19支持使用R语言中的formula形式:
mydf <- data.frame(Entrez=names(geneList), FC=geneList)
mydf <- mydf[abs(mydf$FC) > 1,]
mydf$group <- "upregulated"
mydf$group[mydf$FC < 0] <- "downregulated"
mydf$othergroup <- "A"
mydf$othergroup[abs(mydf$FC) > 2] <- "B"
str(mydf)
## 'data.frame': 1324 obs. of 4 variables:
## $ Entrez : chr "4312" "8318" "10874" "55143" ...
## $ FC : num 4.57 4.51 4.42 4.14 3.88 ...
## $ group : chr "upregulated" "upregulated" "upregulated" "upregulated" ...
## $ othergroup: chr "B" "B" "B" "B" ...
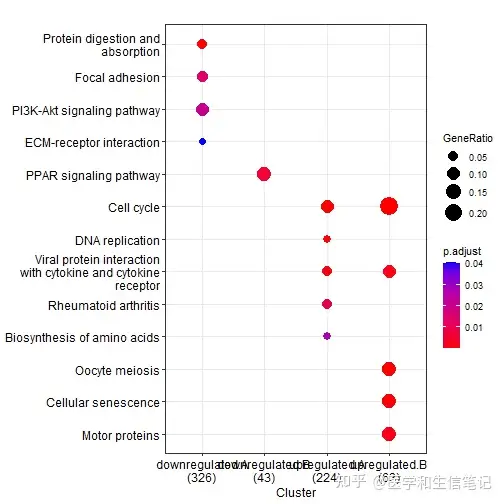
formula_res <- compareCluster(Entrez~group+othergroup, data=mydf, fun="enrichKEGG")
head(formula_res)
## Cluster group othergroup ID
## 1 downregulated.A downregulated A hsa04974
## 2 downregulated.A downregulated A hsa04510
## 3 downregulated.A downregulated A hsa04151
## 4 downregulated.A downregulated A hsa04512
## 5 downregulated.B downregulated B hsa03320
## 6 upregulated.A upregulated A hsa04110
## Description GeneRatio BgRatio pvalue p.adjust
## 1 Protein digestion and absorption 16/326 103/8465 1.731006e-06 4.864127e-04
## 2 Focal adhesion 20/326 203/8465 1.030192e-04 1.447420e-02
## 3 PI3K-Akt signaling pathway 28/326 354/8465 2.240338e-04 2.098450e-02
## 4 ECM-receptor interaction 11/326 89/8465 5.802099e-04 4.075974e-02
## 5 PPAR signaling pathway 5/43 75/8465 3.532307e-05 5.581045e-03
## 6 Cell cycle 21/224 157/8465 7.699196e-10 1.963295e-07
## qvalue
## 1 4.555279e-04
## 2 1.355516e-02
## 3 1.965209e-02
## 4 3.817170e-02
## 5 5.131141e-03
## 6 1.750554e-07
## 省略可视化也是一样的支持:
dotplot(ck)
dotplot(formula_res)
cnetplot(ck)
除了以上的富集分析外,还有几个其他类似方法没介绍到,单基因富集分析、基因集变异分析GSVA、ssGSEA,这些等以后再介绍。
由于输出内容非常多,本文中很多输出都省略了,大家可以自己敲一敲代码看看详细的结果。
对富集结果取子集
富集分析得到的结果都是一个对象,但是可以直接用as.data.frame转化为data.frame,然后你就可以直接保存为CSV格式了。
#以ego为例,其他也是一样
class(ego)
## [1] "enrichResult"
## attr(,"package")
## [1] "DOSE"
go_res <- as.data.frame(ego)
class(go_res)
## [1] "data.frame"
write.csv(go_res,file = "gores.csv",quote = F)所有的富集结果都是可以像这样转化的,方便你进行个性化的操作。
当然,如果你只是单纯的想要对这个结果取子集,也是很简单的,因为:
基本上data.frame取子集那些方法,包括[, [[, >$等操作符,都被我重新定义,也就是说,对于富集分析的结果,你完全可以像data.frame一样的操作。参考:https://mp.weixin.qq.com/s/k-Lbpz8M616Wo0St0TQ-2Q
不过在此之前,我们先探索下富集分析的结果。
dim(ego)
## [1] 153 10
#head(ego)这个富集结果共有153行,10列。其实这个数据就在ego@result这个数据框中,所有的操作也是在对这个数据框进行操作。
如果你不知道这10个列都是哪些信息(即你不知道富集分析可以得到哪些信息),可以直接查看这个结果的列名:
names(ego@result)
## [1] "ONTOLOGY" "ID" "Description" "GeneRatio" "BgRatio" "pvalue"
## [7] "p.adjust" "qvalue" "geneID" "Count" 可以看到,就是常见的富集到的条目,P值等信息,其实各种数据操作和画图等,都是针对这个数据的,只不过clusterprofiler和enrichplot对这个过程进行了简化,不用你提取数据也能直接使用。
取子集非常简单:
#asis=T让子集也是富集分析结果对象,可以直接用
y = ego[ego$pvalue < 0.001, asis=T]
dim(y)
## [1] 153 10
class(y)
## [1] "enrichResult"
## attr(,"package")
## [1] "DOSE"如何查看某个条目下的所有基因名字,很简单,也是不断的取子集操作:
# 第一个条目的所有基因
gsea_res[[gsea_res$ID[[1]]]]
## [1] "1673" "5068" "2919" "5967" "5968" "1670" "1671" "2920" "3553"
## [10] "6373" "4283" "10563" "6374" "6372" "6283" "2921" "6280" "1755"
## [19] "3627" "5266" "718" "725" "4057" "931" "629" "3426" "6278"
## [28] "28461" "1604" "6347" "54210" "3495" "1380" "1378" "1191" "722"
## [37] "1880" "5199" "5648" "5788" "1235" "9308" "717" "3569" "3500"
## [46] "710" "11005" "5196" "716" "715" "1236" "64127" "5079" "940"
## [55] "3507" "28639" "10417" "2219" "5452" "124976" "2213" "5450" "10578"医学和生信笔记公众号主要分享4大块内容:
- 生信数据挖掘
- 医学统计分析
- 机器学习
- 临床预测模型
前期主要是以医学统计和临床预测模型为主,关于生信挖掘和机器学习的内容偏少,所以后面会逐渐增加这方面的内容,除了常见的生信分析外,还会涉及一些SCI图表学习等内容。
富集分析作为了解基因功能的常用方法,也是各种生信相关SCI中的常客,基本上是必会的内容。
今天为大家带来常见的富集分析类型,其实主要就是两类:
ORA(Over-Representation Analysis),通过超几何分布检验实现GSEA(gene set enrichment analysis),基因集富集分析
除此之外,还有ssGSEA、GSVA等,我们等下次再介绍。富集分析的可视化,我们也是放到下次再介绍,因为1条推文根本说不完,太多了!
目前做ORA和GSEA,肯定是首选clusterProfiler了,不管是使用的便捷性,还是对各种富集分析的完美支持,还是对各种最新数据库的支持,clusterProfiler都是最好的选择!
关于各种富集分析的底层原理,我们这里就不说了,大家感兴趣的可以自己学习下。
library(clusterProfiler)
##
## clusterProfiler v4.6.2 For help: https://yulab-smu.top/biomedical-knowledge-mining-book/
##
## If you use clusterProfiler in published research, please cite:
## T Wu, E Hu, S Xu, M Chen, P Guo, Z Dai, T Feng, L Zhou, W Tang, L Zhan, X Fu, S Liu, X Bo, and G Yu. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. The Innovation. 2021, 2(3):100141
##
## Attaching package: 'clusterProfiler'
## The following object is masked from 'package:stats':
##
## filterGO富集分析
clusterProfiler为我们提供了3种GO富集分析:
- GO分类
- GO ORA
- GO GSEA
GO分类
基于特定GO分布对基因进行分类,使用起来超级简单,下面我们用示例数据为大家演示。
data(geneList, package="DOSE")
gene <- names(geneList)[abs(geneList) > 2]
# Entrez gene ID
head(gene)
## [1] "4312" "8318" "10874" "55143" "55388" "991"geneList是一个有序的数值型向量,其名字是基因的ID(这里是ENTREZID)。
除了做GSEA分析需要geneList这种格式外,其他都是直接提供基因ID的向量即可:
ggo <- groupGO(gene = gene,
keyType = "ENTREZID",#任意类型都可,不一定非要ENTREZID,这里指定即可
OrgDb = "org.Hs.eg.db",
ont = "CC",# “BP”, “CC”, “MF”
level = 3,
readable = TRUE
)
##
head(ggo)
## ID Description Count GeneRatio geneID
## GO:0000133 GO:0000133 polarisome 0 0/207
## GO:0000408 GO:0000408 EKC/KEOPS complex 0 0/207
## GO:0000417 GO:0000417 HIR complex 0 0/207
## GO:0000444 GO:0000444 MIS12/MIND type complex 0 0/207
## GO:0000808 GO:0000808 origin recognition complex 0 0/207
## GO:0000930 GO:0000930 gamma-tubulin complex 0 0/207GO过代表分析
这个就是大家最常见的GO富集分析了,使用enrichGO函数实现,各种参数也是和上面差不多:
ego <- enrichGO(gene = gene,
universe = names(geneList),#背景基因
OrgDb = "org.Hs.eg.db",
ont = "ALL",#"BP", "MF", "CC"
pAdjustMethod = "BH",
pvalueCutoff = 0.01,
qvalueCutoff = 0.05,
readable = TRUE)
ego
## #
## # over-representation test
## #
## #...@organism Homo sapiens
## #...@ontology GOALL
## #...@keytype ENTREZID
## #...@gene chr [1:207] "4312" "8318" "10874" "55143" "55388" "991" "6280" "2305" ...
## #...pvalues adjusted by 'BH' with cutoff <0.01
## #...153 enriched terms found
## 'data.frame': 153 obs. of 10 variables:
## $ ONTOLOGY : chr "BP" "BP" "BP" "BP" ...
## $ ID : chr "GO:0140014" "GO:0000280" "GO:0000070" "GO:0048285" ...
## $ Description: chr "mitotic nuclear division" "nuclear division" "mitotic sister chromatid segregation" "organelle fission" ...
## $ GeneRatio : chr "32/195" "36/195" "26/195" "37/195" ...
## $ BgRatio : chr "263/11590" "357/11590" "165/11590" "391/11590" ...
## $ pvalue : num 7.10e-19 1.96e-18 2.60e-18 5.24e-18 6.54e-18 ...
## $ p.adjust : num 2.16e-15 2.65e-15 2.65e-15 3.99e-15 3.99e-15 ...
## $ qvalue : num 2.00e-15 2.44e-15 2.44e-15 3.68e-15 3.68e-15 ...
## $ geneID : chr 省略
head(ego)
## ONTOLOGY ID Description GeneRatio
## GO:0140014 BP GO:0140014 mitotic nuclear division 32/195
## GO:0000280 BP GO:0000280 nuclear division 36/195
## GO:0000070 BP GO:0000070 mitotic sister chromatid segregation 26/195
## GO:0048285 BP GO:0048285 organelle fission 37/195
## GO:0000819 BP GO:0000819 sister chromatid segregation 27/195
## GO:0007059 BP GO:0007059 chromosome segregation 30/195
## BgRatio pvalue p.adjust qvalue
## GO:0140014 263/11590 7.101540e-19 2.163839e-15 1.998149e-15
## GO:0000280 357/11590 1.956617e-18 2.645070e-15 2.442530e-15
## GO:0000070 165/11590 2.604269e-18 2.645070e-15 2.442530e-15
## GO:0048285 391/11590 5.235148e-18 3.987643e-15 3.682300e-15
## GO:0000819 188/11590 6.543556e-18 3.987643e-15 3.682300e-15
## GO:0007059 278/11590 2.850765e-16 1.447713e-13 1.336859e-13
## geneID
## GO:0140014 CDCA8/CDC20/KIF23/CENPE/MYBL2/NDC80/NCAPH/DLGAP5/UBE2C/NUSAP1/TPX2/TACC3/NEK2/UBE2S/MAD2L1/KIF18A/CDT1/BIRC5/KIF11/TTK/NCAPG/AURKB/CHEK1/TRIP13/PRC1/KIFC1/KIF18B/AURKA/CCNB1/KIF4A/PTTG1/BMP4
#省略
## Count
## GO:0140014 32
## GO:0000280 36
## GO:0000070 26
## GO:0048285 37
## GO:0000819 27
## GO:0007059 30GO的数据库条目可能有很多是在解释同一个东西,所clusterProfielr提供了simplify能够去除冗余的GO:
simple_ego <- simplify(ego)目前这个函数支持来自enrichGO、gseGO、compareCluster的结果,非常实用!
GO基因集富集分析
GO版本的GSEA富集分析, gene set enrichment analysis,通过gseGO函数实现:
ego3 <- gseGO(geneList = geneList,
OrgDb = "org.Hs.eg.db",
ont = "CC",
minGSSize = 100,
maxGSSize = 500,# 只有基因集数量在[minGSSize, maxGSSize]之间的才会被检验
pvalueCutoff = 0.05,
verbose = FALSE)
head(ego3)
## ID Description setSize
## GO:0000775 GO:0000775 chromosome, centromeric region 188
## GO:0000779 GO:0000779 condensed chromosome, centromeric region 138
## GO:0000776 GO:0000776 kinetochore 130
## GO:0000228 GO:0000228 nuclear chromosome 175
## GO:0098687 GO:0098687 chromosomal region 305
## GO:0000793 GO:0000793 condensed chromosome 210
## enrichmentScore NES pvalue p.adjust qvalue rank
## GO:0000775 0.5970689 2.652931 1e-10 1.62e-09 1.073684e-09 530
## GO:0000779 0.6216009 2.648130 1e-10 1.62e-09 1.073684e-09 798
## GO:0000776 0.6230073 2.609292 1e-10 1.62e-09 1.073684e-09 449
## GO:0000228 0.5875327 2.580497 1e-10 1.62e-09 1.073684e-09 1905
## GO:0098687 0.5419949 2.545830 1e-10 1.62e-09 1.073684e-09 1721
## GO:0000793 0.5507360 2.468350 1e-10 1.62e-09 1.073684e-09 1721
## leading_edge
## GO:0000775 tags=20%, list=4%, signal=19%
## GO:0000779 tags=24%, list=6%, signal=23%
## GO:0000776 tags=21%, list=4%, signal=20%
## GO:0000228 tags=34%, list=15%, signal=29%
## GO:0098687 tags=27%, list=14%, signal=24%
## GO:0000793 tags=28%, list=14%, signal=24%
## 省略前面说过,做GSEA富集分析对数据格式是有要求的:
- 按照递减排序的数值型向量,
- 向量的名字是gene id
KEGG富集分析
KEGG自2012年起不再免费提供FTP下载服务,但是clusterProfiler包支持使用KEGG网站下载最新的在线版本的KEGG数据,KEGG pathway和module都支持哦。
KEGG作为一个常用的数据库是有官方网站的,大家可以自己去熟悉一下:https://www.kegg.jp/
KEGG pathway ORA
KEGG通路ORA分析,KEGG pathway over-representation analysis,通过enrichKEGG函数实现,这些函数的名字都是有规律的,方便你记忆和使用:
kk <- enrichKEGG(gene = gene,
organism = 'hsa',
pvalueCutoff = 0.05)
## Reading KEGG annotation online: "https://rest.kegg.jp/link/hsa/pathway"...
## Reading KEGG annotation online: "https://rest.kegg.jp/list/pathway/hsa"...
head(kk)
## ID Description
## hsa04110 hsa04110 Cell cycle
## hsa04114 hsa04114 Oocyte meiosis
## hsa04218 hsa04218 Cellular senescence
## hsa04061 hsa04061 Viral protein interaction with cytokine and cytokine receptor
## hsa03320 hsa03320 PPAR signaling pathway
## hsa04814 hsa04814 Motor proteins
## GeneRatio BgRatio pvalue p.adjust qvalue
## hsa04110 15/106 157/8465 8.177242e-10 1.717221e-07 1.695702e-07
## hsa04114 10/106 131/8465 5.049610e-06 5.302091e-04 5.235648e-04
## hsa04218 10/106 156/8465 2.366003e-05 1.639157e-03 1.618617e-03
## hsa04061 8/106 100/8465 3.326461e-05 1.639157e-03 1.618617e-03
## hsa03320 7/106 75/8465 3.902756e-05 1.639157e-03 1.618617e-03
## hsa04814 10/106 193/8465 1.433387e-04 5.016856e-03 4.953988e-03
## geneID
## hsa04110 8318/991/9133/10403/890/983/4085/81620/7272/9212/1111/9319/891/4174/9232
## hsa04114 991/9133/983/4085/51806/6790/891/9232/3708/5241
## hsa04218 2305/4605/9133/890/983/51806/1111/891/776/3708
## hsa04061 3627/10563/6373/4283/6362/6355/9547/1524
## hsa03320 4312/9415/9370/5105/2167/3158/5346
## hsa04814 9493/1062/81930/3832/3833/146909/10112/24137/4629/7802
## Count
## hsa04110 15
## hsa04114 10
## hsa04218 10
## hsa04061 8
## hsa03320 7
## hsa04814 10KEGG pathway GSEA
KEGG通路基因集富集分析,KEGG pathway gene set enrichment analysis,通过gseKEGG实现:
kk2 <- gseKEGG(geneList = geneList,
organism = 'hsa',
minGSSize = 120,
pvalueCutoff = 0.05,
verbose = FALSE)
head(kk2)
## ID Description setSize
## hsa04110 hsa04110 Cell cycle 139
## hsa05169 hsa05169 Epstein-Barr virus infection 193
## hsa04613 hsa04613 Neutrophil extracellular trap formation 130
## hsa05166 hsa05166 Human T-cell leukemia virus 1 infection 202
## hsa04510 hsa04510 Focal adhesion 192
## hsa04218 hsa04218 Cellular senescence 141
## enrichmentScore NES pvalue p.adjust qvalue rank
## hsa04110 0.6637551 2.876742 1.000000e-10 7.900000e-09 5.263158e-09 1155
## hsa05169 0.4335010 1.956648 2.185378e-07 8.632244e-06 5.750996e-06 2820
## hsa04613 0.4496569 1.910465 7.157902e-06 1.884914e-04 1.255772e-04 2575
## hsa05166 0.3893613 1.765888 1.482811e-05 2.928551e-04 1.951067e-04 1955
## hsa04510 -0.4233234 -1.741010 3.210562e-05 5.072688e-04 3.379539e-04 2183
## hsa04218 0.4115945 1.788002 8.805195e-05 1.159351e-03 7.723855e-04 1155
## leading_edge
## hsa04110 tags=36%, list=9%, signal=33%
## hsa05169 tags=39%, list=23%, signal=31%
## hsa04613 tags=37%, list=21%, signal=30%
## hsa05166 tags=26%, list=16%, signal=22%
## hsa04510 tags=28%, list=17%, signal=23%
## hsa04218 tags=17%, list=9%, signal=16%
## core_enrichment
## hsa04110 8318/991/9133/10403/890/983/4085/81620/7272/9212/1111/9319/891/4174/9232/4171/993/990/5347/701/9700/898/23594/4998/9134/4175/4173/10926/6502/994/699/4609/5111/26271/1869/1029/8317/4176/2810/3066/1871/1031/9088/995/1019/4172/5885/11200/7027/1875
#省略KEGG module ORA
KEGG模块ORA分析,KEGG module over-representation analysis,通过enrichMKEGG实现:
mkk <- enrichMKEGG(gene = gene,
organism = 'hsa',
pvalueCutoff = 1,
qvalueCutoff = 1)
## Reading KEGG annotation online: "https://rest.kegg.jp/link/hsa/module"...
## Reading KEGG annotation online: "https://rest.kegg.jp/list/module"...
head(mkk)
## ID
## M00912 M00912
## M00095 M00095
## M00053 M00053
## M00938 M00938
## M00003 M00003
## M00049 M00049
## Description
## M00912 NAD biosynthesis, tryptophan => quinolinate => NAD
## M00095 C5 isoprenoid biosynthesis, mevalonate pathway
## M00053 Deoxyribonucleotide biosynthesis, ADP/GDP/CDP/UDP => dATP/dGTP/dCTP/dUTP
## M00938 Pyrimidine deoxyribonucleotide biosynthesis, UDP => dTTP
## M00003 Gluconeogenesis, oxaloacetate => fructose-6P
## M00049 Adenine ribonucleotide biosynthesis, IMP => ADP,ATP
## GeneRatio BgRatio pvalue p.adjust qvalue geneID Count
## M00912 2/9 12/852 0.00620266 0.03721596 0.03264558 23475/3620 2
## M00095 1/9 10/852 0.10126209 0.18449380 0.16183667 3158 1
## M00053 1/9 11/852 0.11086855 0.18449380 0.16183667 6241 1
## M00938 1/9 14/852 0.13914269 0.18449380 0.16183667 6241 1
## M00003 1/9 18/852 0.17559737 0.18449380 0.16183667 5105 1
## M00049 1/9 19/852 0.18449380 0.18449380 0.16183667 26289 1KEGG module GSEA
KEGG模块GSEA分析,KEGG module gene set enrichment analysis
mkk2 <- gseMKEGG(geneList = geneList,
organism = 'hsa',
pvalueCutoff = 1)
## preparing geneSet collections...
## GSEA analysis...
## leading edge analysis...
## done...
head(mkk2)
## ID Description
## M00001 M00001 Glycolysis (Embden-Meyerhof pathway), glucose => pyruvate
## M00938 M00938 Pyrimidine deoxyribonucleotide biosynthesis, UDP => dTTP
## M00002 M00002 Glycolysis, core module involving three-carbon compounds
## M00035 M00035 Methionine degradation
## M00104 M00104 Bile acid biosynthesis, cholesterol => cholate/chenodeoxycholate
## M00009 M00009 Citrate cycle (TCA cycle, Krebs cycle)
## setSize enrichmentScore NES pvalue p.adjust qvalue rank
## M00001 24 0.5739036 1.746110 0.006607415 0.2180447 0.2086552 2886
## M00938 10 0.6648004 1.550423 0.041882396 0.4607064 0.4408673 648
## M00002 11 0.6421781 1.548820 0.039734634 0.4607064 0.4408673 1381
## M00035 11 0.5824213 1.404697 0.090476190 0.6366300 0.6092153 1555
## M00104 10 -0.5876900 -1.370515 0.108247423 0.6366300 0.6092153 961
## M00009 22 0.4504911 1.340658 0.125858124 0.6366300 0.6092153 3514
## leading_edge
## M00001 tags=54%, list=23%, signal=42%
## M00938 tags=40%, list=5%, signal=38%
## M00002 tags=55%, list=11%, signal=49%
## M00035 tags=45%, list=12%, signal=40%
## M00104 tags=50%, list=8%, signal=46%
## M00009 tags=50%, list=28%, signal=36%
## core_enrichment
## M00001 5214/3101/2821/7167/2597/5230/2023/5223/5315/3099/5232/2027/5211
## M00938 6241/7298/4830/1841
## M00002 7167/2597/5230/2023/5223/5315
## M00035 875/1789/191/1788/1786
## M00104 6342/10998/1581/3295/8309
## M00009 3418/50/4190/3419/2271/3421/55753/3417/1431/6389/4191KEGG通路可视化
执行以下代码会直接打开网页查看:
browseKEGG(kk, 'hsa04110')或者也可以下载到本地,通过pathview这个包实现:
library("pathview")
hsa04110 <- pathview(gene.data = geneList,
pathway.id = "hsa04110",
species = "hsa",
limit = list(gene=max(abs(geneList)), cpd=1))

WikiPathways富集分析
WikiPathways(https://www.wikipathways.org/)和KEGG类似,也是一个通路数据库。

富集分析本质上是对基因注释,以了解这些基因的功能,不同的数据库有不同的侧重点,可能今天说的这些注释数据库有几个你没听过,但是不要紧,你可以自己去了解下即可,如果它们都不符合你的要求,你可以自己定义一个注释数据!
WikiPathways ORA
通过enrichWP函数实现,不过需要良好的网络环境,不然就会报错~
enrichWP(gene, organism = "Homo sapiens")
## #
## # over-representation test
## #
## #...@organism Homo sapiens
## #...@ontology WikiPathways
## #...@keytype ENTREZID
## #...@gene chr [1:207] "4312" "8318" "10874" "55143" "55388" "991" "6280" "2305" ...
## #...pvalues adjusted by 'BH' with cutoff <0.05
## #...8 enriched terms found
## 'data.frame': 8 obs. of 9 variables:
## $ ID : chr "WP2446" "WP2361" "WP179" "WP3942" ...
## $ Description: chr "Retinoblastoma gene in cancer" "Gastric cancer network 1" "Cell cycle" "PPAR signaling pathway" ...
## $ GeneRatio : chr "11/113" "6/113" "10/113" "7/113" ...
## $ BgRatio : chr "90/8444" "28/8444" "120/8444" "68/8444" ...
## $ pvalue : num 2.59e-08 1.49e-06 4.18e-06 3.15e-05 3.70e-05 ...
## $ p.adjust : num 7.14e-06 2.06e-04 3.84e-04 2.04e-03 2.04e-03 ...
## $ qvalue : num 6.42e-06 1.85e-04 3.46e-04 1.84e-03 1.84e-03 ...
## $ geneID : chr "8318/9133/7153/6241/890/983/81620/7272/1111/891/24137" "4605/7153/11065/22974/6286/6790" "8318/991/9133/890/983/7272/1111/891/4174/9232" "4312/9415/9370/5105/2167/3158/5346" ...
## $ Count : int 11 6 10 7 4 7 8 10
## #...Citation
## T Wu, E Hu, S Xu, M Chen, P Guo, Z Dai, T Feng, L Zhou, W Tang, L Zhan, X Fu, S Liu, X Bo, and G Yu.
## clusterProfiler 4.0: A universal enrichment tool for interpreting omics data.
## The Innovation. 2021, 2(3):100141WikiPathways gene set enrichment analysis
通过gseWP实现,也是需要良好的网络环境:
gseWP(geneList, organism = "Homo sapiens")
## preparing geneSet collections...
## GSEA analysis...
## leading edge analysis...
## done...
## #
## # Gene Set Enrichment Analysis
## #
## #...@organism Homo sapiens
## #...@setType WikiPathways
## #...@keytype ENTREZID
## #...@geneList Named num [1:12495] 4.57 4.51 4.42 4.14 3.88 ...
## - attr(*, "names")= chr [1:12495] "4312" "8318" "10874" "55143" ...
## #...nPerm
## #...pvalues adjusted by 'BH' with cutoff <0.05
## #...66 enriched terms found
## 'data.frame': 66 obs. of 11 variables:
## $ ID : chr "WP2446" "WP179" "WP466" "WP2361" ...
## $ Description : chr "Retinoblastoma gene in cancer" "Cell cycle" "DNA replication" "Gastric cancer network 1" ...
## $ setSize : int 84 111 42 23 62 201 77 102 41 61 ...
## $ enrichmentScore: num 0.731 0.663 0.792 0.837 0.665 ...
## $ NES : num 2.86 2.75 2.74 2.55 2.47 ...
## $ pvalue : num 1.00e-10 1.00e-10 1.00e-10 1.41e-09 2.66e-09 ...
## $ p.adjust : num 2.12e-08 2.12e-08 2.12e-08 2.24e-07 2.83e-07 ...
## $ qvalue : num 1.77e-08 1.77e-08 1.77e-08 1.87e-07 2.35e-07 ...
## $ rank : num 1333 1234 1002 854 1111 ...
## $ leading_edge : chr "tags=51%, list=11%, signal=46%" "tags=40%, list=10%, signal=36%" "tags=55%, list=8%, signal=51%" "tags=52%, list=7%, signal=49%" ...
## $ core_enrichment: chr "8318/9133/7153/6241/890/983/81620/7272/1111/891/24137/993/898/4998/10733/9134/4175/4173/6502/5984/994/7298/3015"| __truncated__ "8318/991/9133/890/983/7272/1111/891/4174/9232/4171/993/990/5347/9700/898/23594/4998/9134/4175/4173/10926/6502/9"| __truncated__ "8318/55388/81620/4174/4171/990/23594/4998/4175/4173/10926/5984/5111/51053/8317/5427/23649/4176/5982/5557/5558/4172/5424" "4605/7153/11065/22974/6286/6790/1894/56992/4173/1063/9585/8607" ...
## #...Citation
## T Wu, E Hu, S Xu, M Chen, P Guo, Z Dai, T Feng, L Zhou, W Tang, L Zhan, X Fu, S Liu, X Bo, and G Yu.
## clusterProfiler 4.0: A universal enrichment tool for interpreting omics data.
## The Innovation. 2021, 2(3):100141Reactome富集分析
Reactome(https://reactome.org/)也是一个通路数据库~这个网站自带的可视化功能其实很强大,大家可以去试试。

Reactome富集分析是通过ReactomePA包实现的,注意要先加载R包。
Reactome pathway over-representation analysis
Reactome ORA分析,通过函数enrichPathway实现:
library(ReactomePA)
## ReactomePA v1.42.0 For help: https://yulab-smu.top/biomedical-knowledge-mining-book/
##
## If you use ReactomePA in published research, please cite:
## Guangchuang Yu, Qing-Yu He. ReactomePA: an R/Bioconductor package for reactome pathway analysis and visualization. Molecular BioSystems 2016, 12(2):477-479
x <- enrichPathway(gene=gene,
pvalueCutoff = 0.05,
readable=TRUE)
head(x)
## ID Description GeneRatio
## R-HSA-69620 R-HSA-69620 Cell Cycle Checkpoints 22/143
## R-HSA-2500257 R-HSA-2500257 Resolution of Sister Chromatid Cohesion 15/143
## R-HSA-68877 R-HSA-68877 Mitotic Prometaphase 18/143
## R-HSA-69618 R-HSA-69618 Mitotic Spindle Checkpoint 14/143
## R-HSA-68882 R-HSA-68882 Mitotic Anaphase 18/143
## R-HSA-2555396 R-HSA-2555396 Mitotic Metaphase and Anaphase 18/143
## BgRatio pvalue p.adjust qvalue
## R-HSA-69620 293/10899 2.906024e-11 1.330959e-08 1.095112e-08
## R-HSA-2500257 126/10899 8.316509e-11 1.904481e-08 1.567005e-08
## R-HSA-68877 204/10899 1.539628e-10 2.350499e-08 1.933989e-08
## R-HSA-69618 113/10899 2.130715e-10 2.439669e-08 2.007358e-08
## R-HSA-68882 236/10899 1.657466e-09 1.354116e-07 1.114166e-07
## R-HSA-2555396 237/10899 1.773951e-09 1.354116e-07 1.114166e-07
## 省略Reactome pathway gene set enrichment analysis
Reactome GSEA分析,通过函数gsePathway实现:
y <- gsePathway(geneList,
pvalueCutoff = 0.2,
pAdjustMethod = "BH",
verbose = FALSE)
head(y)
## ID Description setSize
## R-HSA-69620 R-HSA-69620 Cell Cycle Checkpoints 237
## R-HSA-453279 R-HSA-453279 Mitotic G1 phase and G1/S transition 142
## R-HSA-69278 R-HSA-69278 Cell Cycle, Mitotic 457
## R-HSA-69306 R-HSA-69306 DNA Replication 137
## R-HSA-69206 R-HSA-69206 G1/S Transition 124
## R-HSA-69481 R-HSA-69481 G2/M Checkpoints 134
## enrichmentScore NES pvalue p.adjust qvalue rank
## R-HSA-69620 0.6678105 3.064037 1e-10 4.105556e-09 3.064327e-09 1905
## R-HSA-453279 0.7016040 3.001137 1e-10 4.105556e-09 3.064327e-09 1155
## R-HSA-69278 0.6077642 2.974757 1e-10 4.105556e-09 3.064327e-09 1769
## R-HSA-69306 0.6968511 2.961013 1e-10 4.105556e-09 3.064327e-09 1769
## R-HSA-69206 0.7046866 2.949856 1e-10 4.105556e-09 3.064327e-09 1155
## R-HSA-69481 0.6876905 2.925269 1e-10 4.105556e-09 3.064327e-09 1905
## leading_edge
## R-HSA-69620 tags=43%, list=15%, signal=37%
## R-HSA-453279 tags=41%, list=9%, signal=37%
## R-HSA-69278 tags=36%, list=14%, signal=32%
## R-HSA-69306 tags=49%, list=14%, signal=42%
## R-HSA-69206 tags=42%, list=9%, signal=38%
## R-HSA-69481 tags=49%, list=15%, signal=42% core_enrichment
## R-HSA-69620 8318/55143/55388/991/1062/9133/10403/11065/220134/79019/55839/27338/890/983/54821/4085/81930/332/9212/1111/891/4174/4171/11004/993/990/5347/701/51512/9156/11130/79682/57405/2491/898/23594/4998/9134/4175/4173/10926/5984/1058/699/1063/85236/5688/5709/55055/641/1029/5698/1763/8970/5693/8317/4176/5713/79980/5982/9735/5721/2810/5691/9088/995/5685/7468/4172/7336/5690/5684/83990/5686/5695/11200/10213/8345/7534/80010/23198/5983/7979/54908/6396/4683/63967/3018/5699/5714/5702/3014/5905/3619/5708/55166/5692/10393/8290/5704/580/6119
## R-HSA-453279 省略 可视化
这里简单说下Reactome富集分析的可视化,因为viewPathway函数是它专用的:
viewPathway("E2F mediated regulation of DNA replication",
readable = TRUE,
foldChange = geneList)
## Loading required package: graphite

疾病富集分析
大家比较常用的是对基因做富集分析,但是疾病也是可以做富集分析的,有基因本体论(GO),也有疾病本体论(DO)
DOSE包是专门用来做疾病富集分析的。DOSE提供了五种方法来测量DO-term和基因产物之间的语义相似性,也是通过ORA和GSEA将疾病与基因相关联。
library(DOSE)
## DOSE v3.24.2 For help: https://yulab-smu.top/biomedical-knowledge-mining-book/
##
## If you use DOSE in published research, please cite:
## Guangchuang Yu, Li-Gen Wang, Guang-Rong Yan, Qing-Yu He. DOSE: an R/Bioconductor package for Disease Ontology Semantic and Enrichment analysis. Bioinformatics 2015, 31(4):608-609Disease over-representation analysis
DOSE支持疾病本体论(DO)、癌症基因网络和疾病基因网络(DisGeNET)的富集分析。
- Over-representation analysis for disease ontology
疾病本体论(DO)的ORA分析,和GO的ORA分析一模一样:
x <- enrichDO(gene = gene,
ont = "DO",
pvalueCutoff = 0.05,
pAdjustMethod = "BH",
universe = names(geneList),
minGSSize = 10,
maxGSSize = 500,
qvalueCutoff = 0.05,
readable = FALSE)
head(x)
## ID Description GeneRatio BgRatio
## DOID:1107 DOID:1107 esophageal carcinoma 15/157 195/7744
## DOID:5041 DOID:5041 esophageal cancer 15/157 229/7744
## DOID:3459 DOID:3459 breast carcinoma 22/157 461/7744
## DOID:3748 DOID:3748 esophagus squamous cell carcinoma 10/157 131/7744
## DOID:8719 DOID:8719 in situ carcinoma 10/157 132/7744
## DOID:0060071 DOID:0060071 pre-malignant neoplasm 11/157 159/7744
## pvalue p.adjust qvalue
## DOID:1107 8.724913e-06 0.006343012 0.005675786
## DOID:5041 5.832237e-05 0.021200181 0.018970118
## DOID:3459 1.412419e-04 0.033578155 0.030046043
## DOID:3748 3.100853e-04 0.033578155 0.030046043
## DOID:8719 3.297454e-04 0.033578155 0.030046043
## DOID:0060071 3.664508e-04 0.033578155 0.030046043
## geneID
## DOID:1107 4312/6280/3868/6279/8140/890/5918/332/2146/4321/891/2066/3169/8614/11122
## DOID:5041 4312/6280/3868/6279/8140/890/5918/332/2146/4321/891/2066/3169/8614/11122
## DOID:3459 4312/6280/6279/7153/6241/4751/890/4085/332/6286/2146/6790/891/9232/771/4250/2066/3169/10647/5304/5241/10551
## DOID:3748 6280/3868/6279/8140/5918/2146/891/2066/3169/11122
## DOID:8719 6280/7153/6278/10232/332/9212/6790/891/2066/5241
## DOID:0060071 6280/7153/6278/10232/332/9212/4321/6790/891/2066/5241
## Count
## DOID:1107 15
## DOID:5041 15
## DOID:3459 22
## DOID:3748 10
## DOID:8719 10
## DOID:0060071 11注意:ont参数可以是"DO"或"DOLite",DOLite是一个简化后的DO版本,合并了一些冗余的DO term,但是数据不更新了,建议使用ont="DO"
- Over-representation analysis for the network of cancer gene
癌症基因网络的ORA分析:
gene2 <- names(geneList)[abs(geneList) < 3]
head(gene2)
## [1] "8685" "597" "7153" "23397" "6278" "79733"
ncg <- enrichNCG(gene2)
head(ncg)
## ID
## pan-cancer_paediatric pan-cancer_paediatric
## triple_negative_breast_cancer triple_negative_breast_cancer
## bladder_cancer bladder_cancer
## pancreatic_cancer_(all_histologies) pancreatic_cancer_(all_histologies)
## soft_tissue_sarcoma soft_tissue_sarcoma
## paediatric_high-grade_glioma paediatric_high-grade_glioma
## Description
## pan-cancer_paediatric pan-cancer_paediatric
## triple_negative_breast_cancer triple_negative_breast_cancer
## bladder_cancer bladder_cancer
## pancreatic_cancer_(all_histologies) pancreatic_cancer_(all_histologies)
## soft_tissue_sarcoma soft_tissue_sarcoma
## paediatric_high-grade_glioma paediatric_high-grade_glioma
## GeneRatio BgRatio pvalue
## pan-cancer_paediatric 162/2281 183/3177 1.833773e-08
## triple_negative_breast_cancer 71/2281 75/3177 4.290660e-07
## bladder_cancer 97/2281 112/3177 1.253690e-04
## pancreatic_cancer_(all_histologies) 40/2281 42/3177 1.262162e-04
## soft_tissue_sarcoma 26/2281 26/3177 1.742793e-04
## paediatric_high-grade_glioma 25/2281 25/3177 2.434966e-04
## p.adjust qvalue
## pan-cancer_paediatric 1.613721e-06 7.721152e-07
## triple_negative_breast_cancer 1.887890e-05 9.032967e-06
## bladder_cancer 2.776757e-03 1.328592e-03
## pancreatic_cancer_(all_histologies) 2.776757e-03 1.328592e-03
## soft_tissue_sarcoma 3.067315e-03 1.467615e-03
## paediatric_high-grade_glioma 3.073768e-03 1.470702e-03
## geneID
## pan-cancer_paediatric 2146/55353/4609/1029/3575/22806/3418/3066/2120/30012/867/7468/7545/3195/865/64109/4613/613/11177/7490/238/10736/10054/5771/4893/140885/1785/9760/3417/6597/6476/9126/4869/10320/7307/80204/1050/10992/8028/2312/6608/896/894/2196/4849/7023/5093/5079/5293/5727/55181/171017/51322/5781/3718/55294/60/673/8085/5897/4851/1665/51176/1108/7764/10664/6098/2332/2201/6495/3845/7015/1441/2782/64919/4298/23512/8239/29102/6929/8021/6134/6598/4209/5290/22941/8726/207/3717/2033/10716/4928/6932/694/5156/10019/6886/9968/7080/2623/7874/1654/4149/3020/23219/55252/55729/10735/5728/4853/23451/51341/387/3206/6146/79718/2624/63035/3815/171023/23269/25/23592/5896/7403/2260/54880/3716/9203/57178/6777/5789/4297/29072/90/546/120/25836/8289/4345/9611/5925/4763/1997/1499/7157/3399/5295/1387/4602/51564/1027/4005/2322/2078/678/6403/55709/1277/7494/64061/2625
## triple_negative_breast_cancer 省略
## Count
## pan-cancer_paediatric 162
## triple_negative_breast_cancer 71
## bladder_cancer 97
## pancreatic_cancer_(all_histologies) 40
## soft_tissue_sarcoma 26
## paediatric_high-grade_glioma 25- Over-representation analysis for the disease gene network
疾病基因网络的ORA分析,DisGeNET(https://www.disgenet.org/)是一个综合的、全面的基因-疾病数据库,包含基因-疾病关联和snp-基因-疾病关联。
我做网络药理学时经常使用的一个数据库:

疾病-基因关联的富集分析由enrichDGN函数实现,snp-基因-疾病关联的分析由enrichDGNv函数实现。
dgn <- enrichDGN(gene)
head(dgn)
## ID Description GeneRatio BgRatio
## C0007124 C0007124 Noninfiltrating Intraductal Carcinoma 24/204 486/21671
## C3642347 C3642347 Basal-Like Breast Carcinoma 16/204 245/21671
## C1609538 C1609538 Latent Tuberculosis 14/204 183/21671
## C1827293 C1827293 Carcinoma of urinary bladder, invasive 16/204 287/21671
## C0278601 C0278601 Inflammatory Breast Carcinoma 13/204 183/21671
## C4733092 C4733092 estrogen receptor-negative breast cancer 17/204 356/21671
## pvalue p.adjust qvalue
## C0007124 3.806436e-11 1.568251e-07 1.271350e-07
## C3642347 1.555102e-09 3.038993e-06 2.463650e-06
## C1609538 2.212859e-09 3.038993e-06 2.463650e-06
## C1827293 1.491211e-08 1.535947e-05 1.245161e-05
## C0278601 2.050738e-08 1.689808e-05 1.369893e-05
## C4733092 5.070020e-08 3.481414e-05 2.822311e-05
## geneID
## C0007124 4312/6280/6279/7153/6278/9582/55872/3669/4751/5080/2146/1111/3887/6790/9370/125/8839/2066/3169/9547/10647/2625/7021/5241
## C3642347 2305/1062/4605/9833/7368/11065/10232/55765/5163/2146/2568/3620/6790/3169/2625/5241
## C1609538 8685/3627/3669/6373/820/3002/50852/3902/3620/6355/10578/9370/3117/23158
## C1827293 2305/6279/55165/220134/2146/9212/4321/6790/891/8544/9122/23158/652/2625/5241/10551
## C0278601 4312/6278/11065/9582/51203/5918/2146/4321/3887/2066/9547/10647/5241
## C4733092 2305/6278/79733/6241/81930/81620/2146/3620/80129/4137/8839/3169/1408/5304/2625/5241/10551
## Count
## C0007124 24
## C3642347 16
## C1609538 14
## C1827293 16
## C0278601 13
## C4733092 17
snp <- c("rs1401296", "rs9315050", "rs5498", "rs1524668", "rs147377392",
"rs841", "rs909253", "rs7193343", "rs3918232", "rs3760396",
"rs2231137", "rs10947803", "rs17222919", "rs386602276", "rs11053646",
"rs1805192", "rs139564723", "rs2230806", "rs20417", "rs966221")
dgnv <- enrichDGNv(snp)
head(dgnv)
## ID Description GeneRatio BgRatio pvalue
## C0010054 C0010054 Coronary Arteriosclerosis 6/17 440/194515 1.568917e-12
## C0151744 C0151744 Myocardial Ischemia 4/17 103/194515 1.754840e-10
## C0031099 C0031099 Periodontitis 4/17 116/194515 2.839985e-10
## C0007785 C0007785 Cerebral Infarction 4/17 123/194515 3.599531e-10
## C0003850 C0003850 Arteriosclerosis 4/17 267/194515 8.145389e-09
## C0004153 C0004153 Atherosclerosis 4/17 281/194515 9.996713e-09
## p.adjust qvalue
## C0010054 2.761295e-10 NA
## C0151744 1.544259e-08 NA
## C0031099 1.583793e-08 NA
## C0007785 1.583793e-08 NA
## C0003850 2.867177e-07 NA
## C0004153 2.932369e-07 NA
## geneID Count
## C0010054 rs5498/rs147377392/rs11053646/rs1805192/rs2230806/rs20417 6
## C0151744 rs5498/rs147377392/rs11053646/rs1805192 4
## C0031099 rs5498/rs909253/rs1805192/rs20417 4
## C0007785 rs147377392/rs11053646/rs1805192/rs2230806 4
## C0003850 rs5498/rs1805192/rs2230806/rs20417 4
## C0004153 rs5498/rs1805192/rs2230806/rs20417 4Disease gene set enrichment analysis
DO GSEA分析:
y <- gseDO(geneList,
minGSSize = 120,
pvalueCutoff = 0.2,
pAdjustMethod = "BH",
verbose = FALSE)
head(y, 3)
## ID Description setSize
## DOID:399 DOID:399 tuberculosis 181
## DOID:612 DOID:612 primary immunodeficiency disease 305
## DOID:0050338 DOID:0050338 primary bacterial infectious disease 264
## enrichmentScore NES pvalue p.adjust qvalue
## DOID:399 0.5030583 2.252875 1.00000e-10 2.180000e-08 9.789474e-09
## DOID:612 0.4023288 1.898732 4.87282e-09 5.311374e-07 2.385117e-07
## DOID:0050338 0.4150405 1.945371 1.17702e-08 6.414760e-07 2.880602e-07
## rank leading_edge
## DOID:399 2082 tags=40%, list=17%, signal=34%
## DOID:612 2099 tags=33%, list=17%, signal=29%
## DOID:0050338 1864 tags=33%, list=15%, signal=29%
## 省略 癌症基因网络的GSEA分析:
ncg <- gseNCG(geneList,
pvalueCutoff = 0.5,
pAdjustMethod = "BH",
verbose = FALSE)
ncg <- setReadable(ncg, 'org.Hs.eg.db')
head(ncg, 3)
## ID
## pan-gynecological and breast pan-gynecological and breast
## pan-gastric pan-gastric
## esophageal_adenocarcinoma esophageal_adenocarcinoma
## Description setSize
## pan-gynecological and breast pan-gynecological and breast 43
## pan-gastric pan-gastric 49
## esophageal_adenocarcinoma esophageal_adenocarcinoma 92
## enrichmentScore NES pvalue p.adjust
## pan-gynecological and breast -0.5263429 -1.734359 0.004764925 0.1718695
## pan-gastric -0.4993803 -1.683334 0.006365539 0.1718695
## esophageal_adenocarcinoma -0.4235434 -1.550378 0.004980207 0.1718695
## qvalue rank leading_edge
## pan-gynecological and breast 0.1585801 2464 tags=44%, list=20%, signal=36%
## pan-gastric 0.1585801 3280 tags=49%, list=26%, signal=36%
## esophageal_adenocarcinoma 0.1585801 2464 tags=34%, list=20%, signal=27%
## core_enrichment
## pan-gynecological and breast ATM/ZC3H13/NIPBL/SPOP/ARID1A/RASA1/RB1/RNF43/MAP2K4/NF1/CTNNB1/TP53/PIK3R1/CDKN1B/CCND1/ARID5B/MAP3K1/TBX3/GATA3
## pan-gastric BCOR/SOX9/TCF7L2/ATM/CALD1/SEMG2/HTR7/ARID1A/RASA1/RB1/TTBK2/RNF43/CTNNB1/TP53/BCL9/SMAD3/APC/ZFP36L2/TGFBR2/MUC6/MAP3K1/CACNA1C/ATP8B1/CYP4B1
## esophageal_adenocarcinoma NIPBL/ARID1A/RASA1/MSH3/RNF43/CTNNB1/DNAH7/TP53/GATAD1/PIK3R1/STK11/CDKN1B/SPART/APC/SLIT3/LIN7A/SLIT2/TGFBR2/EPHA3/MUC6/ABCB1/NAV3/SYNE1/MAP3K1/SEMA5A/FTO/NUAK1/PCDH9/CHL1/ABCG2/TRIM58疾病基因网络的GSEA分析:
dgn <- gseDGN(geneList,
pvalueCutoff = 0.2,
pAdjustMethod = "BH",
verbose = FALSE)
dgn <- setReadable(dgn, 'org.Hs.eg.db')
head(dgn, 3)
## ID Description setSize enrichmentScore
## C0024266 C0024266 Lymphocytic Choriomeningitis 120 0.5712593
## C4721414 C4721414 Mantle cell lymphoma 368 0.4107437
## C0205682 C0205682 Waist-Hip Ratio 401 -0.4425633
## NES pvalue p.adjust qvalue rank
## C0024266 2.437904 1e-10 2.05275e-07 1.757895e-07 2579
## C4721414 1.951356 1e-10 2.05275e-07 1.757895e-07 1745
## C0205682 -1.946241 1e-10 2.05275e-07 1.757895e-07 2011
## leading_edge
## C0024266 tags=48%, list=21%, signal=38%
## C4721414 tags=26%, list=14%, signal=23%
## C0205682 tags=28%, list=16%, signal=24%
## 省略MeSH富集分析
meshes包支持使用MeSH注释(医学主题词表)对基因列表或整个表达谱进行富集分析(ORA和GSEA)。gendoo/gene2pubmed/RBBH的数据源都支持。
但是进行MeSH富集分析需要通过AnnotationHub下载数据,这一步由于网络问题经常失败...
AnnotationHub在bioconductor 3.16版本(对应R4.2.0版本)之后得到了重用,专门用于存放各种注释数据的R包,通过它可以非常方便的进行查询、下载所有的注释数据!在此之前的版本都是各种注释数据都有自己的专属R包,不管R包多大,我们都可以通过更换镜像实现快速下载!
现在统一之后确实方便大家查找,但是我觉得这样做让大陆用户很为难......基本上都会卡在下载数据这一步(因为这个东西没有镜像可以用了...),即使通过其他方式下载了数据,但是不同的注释数据还有不同的处理方法。
MeSH over-representation analysis
首先自己构建下数据库,AnnotationHub下载数据太慢了...(但是如果网络环境好也是嗖嗖嗖地下载)
# 需要bioc版本在3.14以上
library(AnnotationHub)
library(MeSHDbi)
ah <- AnnotationHub() #第一次运行会问你要不要建立本地缓存,选yes
hsa <- query(ah, c("MeSHDb", "Homo sapiens"))
file_hsa <- hsa[[1]] # 这一步是下载真正的注释,还好不用每次都下载
#file_hsa <- "C:/Users/liyue/AppData/Local/R/cache/R/AnnotationHub/mesh_hs_db_98354"#这个是我自己通过迅雷下载的,可以读取,但是不知道为啥富集分析不能用
db <- MeSHDbi::MeSHDb(file_hsa)来自官方的提示:
MeSH-related packages (MeSH.XXX.eg.db, MeSH.db, MeSH.AOR.db, and MeSH.PCR.db) are deprecated since Bioconductor 3.14. Use AnnotationHub instead. For details, check the vignette of MeSHDbi
以前是可以安装各种注释R包的,换个镜像即可嗖嗖的下载,好日子一去不复返了!
## BioC 2.14 to BioC 3.13 ##
library(MeSH.Hsa.eg.db)
db <- MeSH.Hsa.eg.db接下来就是进行MeSH的ORA分析:
library(meshes)
## meshes v1.24.0
##
## If you use meshes in published research, please cite the most appropriate paper(s):
##
## Guangchuang Yu. Using meshes for MeSH term enrichment and semantic analyses. Bioinformatics 2018, 34(21):3766-3767, doi:10.1093/bioinformatics/bty410
##
## Attaching package: 'meshes'
## The following object is masked from 'package:DOSE':
##
## geneSim
data(geneList, package="DOSE")
de <- names(geneList)[1:100]
x <- enrichMeSH(de, MeSHDb = db, database='gendoo', category = 'C')
## Error in h(simpleError(msg, call)): error in evaluating the argument 'x' in selecting a method for function 'select': object 'db' not found
head(x)
## ID Description GeneRatio BgRatio
## DOID:1107 DOID:1107 esophageal carcinoma 15/157 195/7744
## DOID:5041 DOID:5041 esophageal cancer 15/157 229/7744
## DOID:3459 DOID:3459 breast carcinoma 22/157 461/7744
## DOID:3748 DOID:3748 esophagus squamous cell carcinoma 10/157 131/7744
## DOID:8719 DOID:8719 in situ carcinoma 10/157 132/7744
## DOID:0060071 DOID:0060071 pre-malignant neoplasm 11/157 159/7744
## pvalue p.adjust qvalue
## DOID:1107 8.724913e-06 0.006343012 0.005675786
## DOID:5041 5.832237e-05 0.021200181 0.018970118
## DOID:3459 1.412419e-04 0.033578155 0.030046043
## DOID:3748 3.100853e-04 0.033578155 0.030046043
## DOID:8719 3.297454e-04 0.033578155 0.030046043
## DOID:0060071 3.664508e-04 0.033578155 0.030046043
## geneID
## DOID:1107 4312/6280/3868/6279/8140/890/5918/332/2146/4321/891/2066/3169/8614/11122
## DOID:5041 4312/6280/3868/6279/8140/890/5918/332/2146/4321/891/2066/3169/8614/11122
## DOID:3459 4312/6280/6279/7153/6241/4751/890/4085/332/6286/2146/6790/891/9232/771/4250/2066/3169/10647/5304/5241/10551
## DOID:3748 6280/3868/6279/8140/5918/2146/891/2066/3169/11122
## DOID:8719 6280/7153/6278/10232/332/9212/6790/891/2066/5241
## DOID:0060071 6280/7153/6278/10232/332/9212/4321/6790/891/2066/5241
## Count
## DOID:1107 15
## DOID:5041 15
## DOID:3459 22
## DOID:3748 10
## DOID:8719 10
## DOID:0060071 11MeSH gene set enrichment analysis
然后是GSEA分析:
y <- gseMeSH(geneList, MeSHDb = db, database = 'gene2pubmed', category = "G")
## Error in h(simpleError(msg, call)): error in evaluating the argument 'x' in selecting a method for function 'select': object 'db' not found
head(y)
## ID Description
## DOID:399 DOID:399 tuberculosis
## DOID:612 DOID:612 primary immunodeficiency disease
## DOID:0050338 DOID:0050338 primary bacterial infectious disease
## DOID:104 DOID:104 bacterial infectious disease
## DOID:526 DOID:526 human immunodeficiency virus infectious disease
## DOID:2237 DOID:2237 hepatitis
## setSize enrichmentScore NES pvalue p.adjust
## DOID:399 181 0.5030583 2.252875 1.000000e-10 2.180000e-08
## DOID:612 305 0.4023288 1.898732 4.872820e-09 5.311374e-07
## DOID:0050338 264 0.4150405 1.945371 1.177020e-08 6.414760e-07
## DOID:104 321 0.3899673 1.856122 9.801772e-09 6.414760e-07
## DOID:526 201 0.4147782 1.874250 4.756033e-07 2.073630e-05
## DOID:2237 406 0.3328749 1.613797 1.567985e-06 5.697013e-05
## qvalue rank leading_edge
## DOID:399 9.789474e-09 2082 tags=40%, list=17%, signal=34%
## DOID:612 2.385117e-07 2099 tags=33%, list=17%, signal=29%
## DOID:0050338 2.880602e-07 1864 tags=33%, list=15%, signal=29%
## DOID:104 2.880602e-07 2330 tags=36%, list=19%, signal=30%
## DOID:526 9.311812e-06 2099 tags=36%, list=17%, signal=31%
## DOID:2237 2.558292e-05 2082 tags=27%, list=17%, signal=23%
## 省略自定义富集分析
有些用户需要自定义自己的注释数据库或者没有支持的物种,也可以自己DIY。
clusterProfiler提供了用于超几何检验的enricher()函数和用于基因集富集分析的GSEA()函数,可以让用户根据自己的注释信息进行富集分析。
需要两个额外的参数TERM2GENE和TERM2NAME。TERM2GENE是一个数据框,第一列是term-ID,第二列是相应的映射基因;TERM2NAME也是一个数据框,第一列是term-ID,第二列是相应的术语名称。其中TERM2NAME是可选的。
MiSigdb
下面是一个用MiSigdb数据库进行富集分析的例子。MiSigdb可以非常方便的进行查询和下载MiSigdb数据库的基因集。
查看支持的物种:
library(msigdbr)
msigdbr_show_species()
## [1] "Anolis carolinensis" "Bos taurus"
## [3] "Caenorhabditis elegans" "Canis lupus familiaris"
## [5] "Danio rerio" "Drosophila melanogaster"
## [7] "Equus caballus" "Felis catus"
## [9] "Gallus gallus" "Homo sapiens"
## [11] "Macaca mulatta" "Monodelphis domestica"
## [13] "Mus musculus" "Ornithorhynchus anatinus"
## [15] "Pan troglodytes" "Rattus norvegicus"
## [17] "Saccharomyces cerevisiae" "Schizosaccharomyces pombe 972h-"
## [19] "Sus scrofa" "Xenopus tropicalis"选择我们想要的数据下载:
m_df <- msigdbr(species = "Homo sapiens")#下载数据
head(m_df, 2) %>% as.data.frame
## gs_cat gs_subcat gs_name gene_symbol entrez_gene ensembl_gene
## 1 C3 MIR:MIR_Legacy AAACCAC_MIR140 ABCC4 10257 ENSG00000125257
## 2 C3 MIR:MIR_Legacy AAACCAC_MIR140 ABRAXAS2 23172 ENSG00000165660
## human_gene_symbol human_entrez_gene human_ensembl_gene gs_id gs_pmid
## 1 ABCC4 10257 ENSG00000125257 M12609
## 2 ABRAXAS2 23172 ENSG00000165660 M12609
## gs_geoid gs_exact_source gs_url
## 1
## 2
## gs_description
## 1 Genes having at least one occurence of the motif AAACCAC in their 3' untranslated region. The motif represents putative target (that is, seed match) of human mature miRNA hsa-miR-140 (v7.1 miRBase).
## 2 Genes having at least one occurence of the motif AAACCAC in their 3' untranslated region. The motif represents putative target (that is, seed match) of human mature miRNA hsa-miR-140 (v7.1 miRBase).我们使用人类的C6注释集:
m_t2g <- msigdbr(species = "Homo sapiens", category = "C6") %>%
dplyr::select(gs_name, entrez_gene)
head(m_t2g)
## # A tibble: 6 × 2
## gs_name entrez_gene
## <chr> <int>
## 1 AKT_UP.V1_DN 57007
## 2 AKT_UP.V1_DN 22859
## 3 AKT_UP.V1_DN 22859
## 4 AKT_UP.V1_DN 137872
## 5 AKT_UP.V1_DN 249
## 6 AKT_UP.V1_DN 271有了这个数据之后,我们就可以根据这个自定义的注释信息做富集分析了:
首先是ORA分析:
em <- enricher(gene, TERM2GENE=m_t2g)
head(em)
## ID Description GeneRatio
## RPS14_DN.V1_DN RPS14_DN.V1_DN RPS14_DN.V1_DN 22/183
## GCNP_SHH_UP_LATE.V1_UP GCNP_SHH_UP_LATE.V1_UP GCNP_SHH_UP_LATE.V1_UP 16/183
## PRC2_EZH2_UP.V1_DN PRC2_EZH2_UP.V1_DN PRC2_EZH2_UP.V1_DN 15/183
## VEGF_A_UP.V1_DN VEGF_A_UP.V1_DN VEGF_A_UP.V1_DN 15/183
## RB_P107_DN.V1_UP RB_P107_DN.V1_UP RB_P107_DN.V1_UP 10/183
## E2F1_UP.V1_UP E2F1_UP.V1_UP E2F1_UP.V1_UP 12/183
## BgRatio pvalue p.adjust qvalue
## RPS14_DN.V1_DN 186/10922 4.657342e-13 7.870908e-11 6.422230e-11
## GCNP_SHH_UP_LATE.V1_UP 181/10922 5.765032e-08 4.871452e-06 3.974838e-06
## PRC2_EZH2_UP.V1_DN 192/10922 7.574546e-07 3.419660e-05 2.790255e-05
## VEGF_A_UP.V1_DN 193/10922 8.093869e-07 3.419660e-05 2.790255e-05
## RB_P107_DN.V1_UP 130/10922 6.394609e-05 2.013355e-03 1.642787e-03
## E2F1_UP.V1_UP 188/10922 7.477287e-05 2.013355e-03 1.642787e-03
## geneID
## RPS14_DN.V1_DN 10874/55388/991/9493/1062/4605/9133/23397/79733/9787/55872/83461/54821/51659/9319/9055/10112/4174/5105/2532/7021/79901
## GCNP_SHH_UP_LATE.V1_UP 55388/7153/79733/6241/9787/51203/983/9212/1111/9319/9055/3833/6790/4174/3169/1580
## PRC2_EZH2_UP.V1_DN 8318/55388/4605/23397/9787/55355/10460/6362/81620/2146/7272/9212/11182/3887/24137
## VEGF_A_UP.V1_DN 8318/9493/1062/9133/10403/6241/9787/4085/332/3832/7272/891/23362/2167/10234
## RB_P107_DN.V1_UP 8318/23397/79733/6241/4085/8208/9055/24137/4174/1307
## E2F1_UP.V1_UP 55388/7153/23397/79733/9787/2146/2842/9212/8208/1111/9055/3833
## Count
## RPS14_DN.V1_DN 22
## GCNP_SHH_UP_LATE.V1_UP 16
## PRC2_EZH2_UP.V1_DN 15
## VEGF_A_UP.V1_DN 15
## RB_P107_DN.V1_UP 10
## E2F1_UP.V1_UP 12再试一下C3基因集:
C3_t2g <- msigdbr(species = "Homo sapiens", category = "C3") %>%
dplyr::select(gs_name, entrez_gene)
head(C3_t2g)
## # A tibble: 6 × 2
## gs_name entrez_gene
## <chr> <int>
## 1 AAACCAC_MIR140 10257
## 2 AAACCAC_MIR140 23172
## 3 AAACCAC_MIR140 81
## 4 AAACCAC_MIR140 81
## 5 AAACCAC_MIR140 90
## 6 AAACCAC_MIR140 8754然后是GSEA分析:
em2 <- GSEA(geneList, TERM2GENE = C3_t2g)
## preparing geneSet collections...
## GSEA analysis...
## leading edge analysis...
## done...
head(em2)
## ID Description setSize
## HSD17B8_TARGET_GENES HSD17B8_TARGET_GENES HSD17B8_TARGET_GENES 401
## E2F1_Q3 E2F1_Q3 E2F1_Q3 187
## E2F1_Q6 E2F1_Q6 E2F1_Q6 185
## SGCGSSAAA_E2F1DP2_01 SGCGSSAAA_E2F1DP2_01 SGCGSSAAA_E2F1DP2_01 129
## E2F_Q4 E2F_Q4 E2F_Q4 193
## E2F1DP1_01 E2F1DP1_01 E2F1DP1_01 184
## enrichmentScore NES pvalue p.adjust
## HSD17B8_TARGET_GENES 0.6368099 3.107103 1.000000e-10 1.655000e-07
## E2F1_Q3 0.4983913 2.220028 1.000000e-10 1.655000e-07
## E2F1_Q6 0.4906657 2.184070 2.011165e-10 2.218985e-07
## SGCGSSAAA_E2F1DP2_01 0.5408182 2.289484 1.032436e-09 8.543410e-07
## E2F_Q4 0.4685394 2.099303 2.297950e-09 9.507767e-07
## E2F1DP1_01 0.4675989 2.083101 2.123251e-09 9.507767e-07
## qvalue rank leading_edge
## HSD17B8_TARGET_GENES 1.310526e-07 1042 tags=36%, list=8%, signal=34%
## E2F1_Q3 1.310526e-07 2045 tags=36%, list=16%, signal=30%
## E2F1_Q6 1.757123e-07 1819 tags=34%, list=15%, signal=30%
## SGCGSSAAA_E2F1DP2_01 6.765174e-07 1797 tags=38%, list=14%, signal=33%
## E2F_Q4 7.528809e-07 1663 tags=29%, list=13%, signal=26%
## E2F1DP1_01 7.528809e-07 1797 tags=33%, list=14%, signal=28%
#省略cellmarker
除了msigdb数据库,还有一个cellmarker数据库,做单细胞的一定见过,我们先下载它的注释数据:
cell_marker_data <- vroom::vroom('http://xteam.xbio.top/CellMarker/download/Human_cell_markers.txt')
## Rows: 2868 Columns: 15
## ── Column specification ─────────────────────────────
## Delimiter: "\t"
## chr (15): speciesType, tissueType, UberonOntologyID, cancerType, cellType, c...
##
## ℹ Use `spec()` to retrieve the full column specification for this data.
## ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
## instead of `cellName`, users can use other features (e.g. `cancerType`)
cells <- cell_marker_data %>%
dplyr::select(cellName, geneID) %>%
dplyr::mutate(geneID = strsplit(geneID, ', ')) %>%
tidyr::unnest()有了数据就可以使用通用的enricher函数进行ORA富集分析了:
x <- enricher(gene, TERM2GENE = cells)
head(x)
## ID Description GeneRatio
## Neural progenitor cell Neural progenitor cell Neural progenitor cell 33/153
## MKI67+ progenitor cell MKI67+ progenitor cell MKI67+ progenitor cell 20/153
## DCLK1+ progenitor cell DCLK1+ progenitor cell DCLK1+ progenitor cell 7/153
## BgRatio pvalue p.adjust qvalue
## Neural progenitor cell 176/11582 3.395466e-29 3.259648e-27 2.716373e-27
## MKI67+ progenitor cell 111/11582 1.426062e-17 6.845100e-16 5.704250e-16
## DCLK1+ progenitor cell 99/11582 3.298755e-04 1.055601e-02 8.796679e-03
## geneID
## Neural progenitor cell 991/9493/9833/9133/10403/7153/259266/6241/9787/11065/55355/55872/51203/83461/22974/10460/79019/55839/890/983/4085/5080/332/3832/9212/51659/9055/891/4174/9232/1602/2018/4137
## MKI67+ progenitor cell 991/9133/10403/7153/259266/6241/9787/11065/51203/22974/10460/890/983/332/9212/9055/891/9232/2167/51705
## DCLK1+ progenitor cell 1307/730/54829/10234/79901/57758/4969
## Count
## Neural progenitor cell 33
## MKI67+ progenitor cell 20
## DCLK1+ progenitor cell 7使用GSEA进行基因集富集分析:
y <- GSEA(geneList, TERM2GENE = cells)
## preparing geneSet collections...
## GSEA analysis...
## leading edge analysis...
## done...
head(y)
## ID
## Neural progenitor cell Neural progenitor cell
## FOXN4+ cell FOXN4+ cell
## Paneth cell Paneth cell
## Leydig precursor cell Leydig precursor cell
## FGFR1HighNME5- epithelial cell FGFR1HighNME5- epithelial cell
## Ciliated epithelial cell Ciliated epithelial cell
## Description setSize
## Neural progenitor cell Neural progenitor cell 149
## FOXN4+ cell FOXN4+ cell 63
## Paneth cell Paneth cell 261
## Leydig precursor cell Leydig precursor cell 184
## FGFR1HighNME5- epithelial cell FGFR1HighNME5- epithelial cell 131
## Ciliated epithelial cell Ciliated epithelial cell 217
## enrichmentScore NES pvalue
## Neural progenitor cell 0.7160571 3.085727 1.000000e-10
## FOXN4+ cell 0.7136090 2.685258 1.000000e-10
## Paneth cell 0.5381280 2.489150 1.000000e-10
## Leydig precursor cell -0.5710583 -2.308894 1.000000e-10
## FGFR1HighNME5- epithelial cell -0.5739472 -2.229562 1.476993e-10
## Ciliated epithelial cell -0.5012031 -2.064339 3.336445e-10
## p.adjust qvalue rank
## Neural progenitor cell 4.125000e-09 2.315789e-09 684
## FOXN4+ cell 4.125000e-09 2.315789e-09 1419
## Paneth cell 4.125000e-09 2.315789e-09 2187
## Leydig precursor cell 4.125000e-09 2.315789e-09 2609
## FGFR1HighNME5- epithelial cell 4.874076e-09 2.736323e-09 1404
## Ciliated epithelial cell 9.175223e-09 5.151002e-09 1440
## leading_edge
## Neural progenitor cell tags=47%, list=5%, signal=45%
## FOXN4+ cell tags=51%, list=11%, signal=45%
## Paneth cell tags=45%, list=18%, signal=38%
## Leydig precursor cell tags=45%, list=21%, signal=36%
## FGFR1HighNME5- epithelial cell tags=39%, list=11%, signal=35%
## Ciliated epithelial cell tags=29%, list=12%, signal=27%
#省略DO, GO, Reactome的分析是有readable参数的,其实是根据OrgDb进行转换的,这和setReadable函数的原理一样,所以用到OrgDb的都有这个参数。
多个基因集富集结果比较
同时对多个基因集进行富集分析,这个功能在单细胞里面简直是绝配!
data(gcSample)
str(gcSample)
## List of 8
## $ X1: chr [1:216] "4597" "7111" "5266" "2175" ...
## $ X2: chr [1:805] "23450" "5160" "7126" "26118" ...
## $ X3: chr [1:392] "894" "7057" "22906" "3339" ...
## $ X4: chr [1:838] "5573" "7453" "5245" "23450" ...
## $ X5: chr [1:929] "5982" "7318" "6352" "2101" ...
## $ X6: chr [1:585] "5337" "9295" "4035" "811" ...
## $ X7: chr [1:582] "2621" "2665" "5690" "3608" ...
## $ X8: chr [1:237] "2665" "4735" "1327" "3192" ...同时对这8个基因集分别进行富集分析:
ck <- compareCluster(geneCluster = gcSample, fun = enrichKEGG)
ck <- setReadable(ck, OrgDb = org.Hs.eg.db, keyType="ENTREZID")
head(ck)
## Cluster ID Description GeneRatio BgRatio
## 1 X2 hsa05169 Epstein-Barr virus infection 23/401 202/8465
## 2 X2 hsa05340 Primary immunodeficiency 8/401 38/8465
## 3 X2 hsa04110 Cell cycle 18/401 157/8465
## 4 X3 hsa04512 ECM-receptor interaction 9/190 89/8465
## 5 X3 hsa04060 Cytokine-cytokine receptor interaction 17/190 297/8465
## 6 X3 hsa04151 PI3K-Akt signaling pathway 19/190 354/8465
## pvalue p.adjust qvalue
## 1 7.981195e-05 0.02466189 0.02310346
## 2 3.300857e-04 0.04463551 0.04181490
## 3 4.333545e-04 0.04463551 0.04181490
## 4 1.617209e-04 0.03272190 0.03037690
## 5 3.565676e-04 0.03272190 0.03037690
## 6 3.622350e-04 0.03272190 0.03037690
## geneID
## 1 LYN/ICAM1/TNFAIP3/E2F1/CCNA2/E2F3/BAK1/RUNX3/BID/IKBKE/TAP2/FAS/CCNE2/RELB/CD3E/ENTPD3/SYK/TRAF3/IKBKB/CD247/NEDD4/CD40/STAT1
## 2 ADA/TAP2/LCK/CD79A/CD3E/CD8A/CD40/DCLRE1C
## 3 CDC20/E2F1/CCNA2/E2F3/BUB1B/CDC6/DBF4/PKMYT1/CDC7/ESPL1/CCNE2/CDKN2A/SFN/BUB1/CHEK2/ORC6/CDC14B/MCM4
## 4 THBS1/HSPG2/COL9A3/ITGB7/CHAD/ITGA7/LAMA4/ITGB8/ITGB5
## 5 CXCL1/TNFRSF11B/LIFR/XCL1/GDF5/TNFRSF17/TNFRSF11A/IL5RA/EPOR/CSF2RA/TNFRSF25/BMP7/BMP4/GDF11/IL21R/IL23A/TGFB2
## 6 CCND2/THBS1/STK11/FGF2/COL9A3/ITGB7/FGF7/ERBB4/CHAD/FGF18/CDK6/PCK1/ITGA7/HGF/EPOR/LAMA4/IKBKB/ITGB8/ITGB5
## Count
## 1 23
## 2 8
## 3 18
## 4 9
## 5 17
## 6 19支持使用R语言中的formula形式:
mydf <- data.frame(Entrez=names(geneList), FC=geneList)
mydf <- mydf[abs(mydf$FC) > 1,]
mydf$group <- "upregulated"
mydf$group[mydf$FC < 0] <- "downregulated"
mydf$othergroup <- "A"
mydf$othergroup[abs(mydf$FC) > 2] <- "B"
str(mydf)
## 'data.frame': 1324 obs. of 4 variables:
## $ Entrez : chr "4312" "8318" "10874" "55143" ...
## $ FC : num 4.57 4.51 4.42 4.14 3.88 ...
## $ group : chr "upregulated" "upregulated" "upregulated" "upregulated" ...
## $ othergroup: chr "B" "B" "B" "B" ...
formula_res <- compareCluster(Entrez~group+othergroup, data=mydf, fun="enrichKEGG")
head(formula_res)
## Cluster group othergroup ID
## 1 downregulated.A downregulated A hsa04974
## 2 downregulated.A downregulated A hsa04510
## 3 downregulated.A downregulated A hsa04151
## 4 downregulated.A downregulated A hsa04512
## 5 downregulated.B downregulated B hsa03320
## 6 upregulated.A upregulated A hsa04110
## Description GeneRatio BgRatio pvalue p.adjust
## 1 Protein digestion and absorption 16/326 103/8465 1.731006e-06 4.864127e-04
## 2 Focal adhesion 20/326 203/8465 1.030192e-04 1.447420e-02
## 3 PI3K-Akt signaling pathway 28/326 354/8465 2.240338e-04 2.098450e-02
## 4 ECM-receptor interaction 11/326 89/8465 5.802099e-04 4.075974e-02
## 5 PPAR signaling pathway 5/43 75/8465 3.532307e-05 5.581045e-03
## 6 Cell cycle 21/224 157/8465 7.699196e-10 1.963295e-07
## qvalue
## 1 4.555279e-04
## 2 1.355516e-02
## 3 1.965209e-02
## 4 3.817170e-02
## 5 5.131141e-03
## 6 1.750554e-07
## 省略可视化也是一样的支持:
dotplot(ck)

dotplot(formula_res)

cnetplot(ck)

除了以上的富集分析外,还有几个其他类似方法没介绍到,单基因富集分析、基因集变异分析GSVA、ssGSEA,这些等以后再介绍。
由于输出内容非常多,本文中很多输出都省略了,大家可以自己敲一敲代码看看详细的结果。
对富集结果取子集
富集分析得到的结果都是一个对象,但是可以直接用as.data.frame转化为data.frame,然后你就可以直接保存为CSV格式了。
#以ego为例,其他也是一样
class(ego)
## [1] "enrichResult"
## attr(,"package")
## [1] "DOSE"
go_res <- as.data.frame(ego)
class(go_res)
## [1] "data.frame"
write.csv(go_res,file = "gores.csv",quote = F)所有的富集结果都是可以像这样转化的,方便你进行个性化的操作。
当然,如果你只是单纯的想要对这个结果取子集,也是很简单的,因为:
基本上data.frame取子集那些方法,包括[, [[, >$等操作符,都被我重新定义,也就是说,对于富集分析的结果,你完全可以像data.frame一样的操作。参考:https://mp.weixin.qq.com/s/k-Lbpz8M616Wo0St0TQ-2Q
不过在此之前,我们先探索下富集分析的结果。
dim(ego)
## [1] 153 10
#head(ego)这个富集结果共有153行,10列。其实这个数据就在ego@result这个数据框中,所有的操作也是在对这个数据框进行操作。
如果你不知道这10个列都是哪些信息(即你不知道富集分析可以得到哪些信息),可以直接查看这个结果的列名:
names(ego@result)
## [1] "ONTOLOGY" "ID" "Description" "GeneRatio" "BgRatio" "pvalue"
## [7] "p.adjust" "qvalue" "geneID" "Count" 可以看到,就是常见的富集到的条目,P值等信息,其实各种数据操作和画图等,都是针对这个数据的,只不过clusterprofiler和enrichplot对这个过程进行了简化,不用你提取数据也能直接使用。
取子集非常简单:
#asis=T让子集也是富集分析结果对象,可以直接用
y = ego[ego$pvalue < 0.001, asis=T]
dim(y)
## [1] 153 10
class(y)
## [1] "enrichResult"
## attr(,"package")
## [1] "DOSE"如何查看某个条目下的所有基因名字,很简单,也是不断的取子集操作:
# 第一个条目的所有基因
gsea_res[[gsea_res$ID[[1]]]]
## [1] "1673" "5068" "2919" "5967" "5968" "1670" "1671" "2920" "3553"
## [10] "6373" "4283" "10563" "6374" "6372" "6283" "2921" "6280" "1755"
## [19] "3627" "5266" "718" "725" "4057" "931" "629" "3426" "6278"
## [28] "28461" "1604" "6347" "54210" "3495" "1380" "1378" "1191" "722"
## [37] "1880" "5199" "5648" "5788" "1235" "9308" "717" "3569" "3500"
## [46] "710" "11005" "5196" "716" "715" "1236" "64127" "5079" "940"
## [55] "3507" "28639" "10417" "2219" "5452" "124976" "2213" "5450" "10578"
## [64] "28912" "3514" "6590" "3119" "6036" "3853" "7124" "4068" "5919"
## [73] "4049" "6406" "8547" "3605" "5473" "3458" "729230" "6480" "240"
## [82] "3123" "4239" 除此之外,dplyr中的很多函数,包括filetr/arrange/select/mutate/slice/summarise,也被重新定义了,支持直接对这些富集结果进行操作,简直不要太方便。
y <- ego %>%
filter(p.adjust < 10^-8) %>%
arrange(Count)
dim(y)
## [1] 27 10
#head(y)
#结果依然是对象!
class(y)
## [1] "enrichResult"
## attr(,"package")
## [1] "DOSE"其他的就不多演示了,大家发挥自己的想象力即可,无非就是对数据框进行操作~
clusterprofiler的富集结果都是一样使用,所以其他的富集分析结果就不再一一演示了。
参考资料
关于clusterprofiler包的用法,y叔专门写了一本书:https://yulab-smu.top/biomedical-knowledge-mining-book/
除此之外,在y叔的公众号也有大量关于clusterprofiler包的推文,我这里直接把合集链接放这里:clusterprofiler合集,其中很多推文是对使用者问题的回答,很多推文中说过问题/bug等,现在基本都被修复了。
## [64] "28912" "3514" "6590" "3119" "6036" "3853" "7124" "4068" "5919" ## [73] "4049" "6406" "8547" "3605" "5473" "3458" "729230" "6480" "240" ## [82] "3123" "4239" 除此之外,dplyr中的很多函数,包括filetr/arrange/select/mutate/slice/summarise,也被重新定义了,支持直接对这些富集结果进行操作,简直不要太方便。
y <- ego %>%
filter(p.adjust < 10^-8) %>%
arrange(Count)
dim(y)
## [1] 27 10
#head(y)
#结果依然是对象!
class(y)
## [1] "enrichResult"
## attr(,"package")
## [1] "DOSE"其他的就不多演示了,大家发挥自己的想象力即可,无非就是对数据框进行操作~
clusterprofiler的富集结果都是一样使用,所以其他的富集分析结果就不再一一演示了。
参考资料
关于clusterprofiler包的用法,y叔专门写了一本书:https://yulab-smu.top/biomedical-knowledge-mining-book/
除此之外,在y叔的公众号也有大量关于clusterprofiler包的推文,我这里直接把合集链接放这里:clusterprofiler合集,其中很多推文是对使用者问题的回答,很多推文中说过问题/bug等,现在基本都被修复了。