REF https://daler.github.io/pybedtools/search.html?q=cat
通过pip 安装
pip install pybedtools
由文件创建BedTool对象
test = pybedtools.BedTool('test.bed')
pybedtools.BedTool( ).sort().merge()
查看前几行数据
>>> a.head()
chr1 1 100 feature1 0 +
chr1 100 200 feature2 0 +
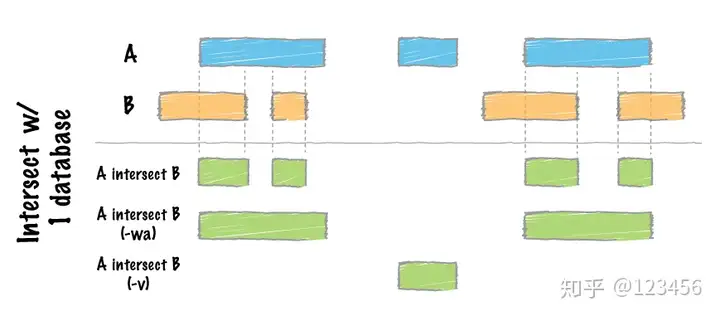
intersect 取交集,
不同参数结果输出不同的区域,A和B是输入的两个BED文件。不输入参数输出结果为两个BED文件的交集,-wa保留和b文件有重叠的全部A文件区域,-v 输出结果为-wa的补集,即B中没有A重叠的区域。可以用图表示为:
a = pybedtools.example_bedtool('a.bed')
b = pybedtools.example_bedtool('b.bed')
a_and_b = a.intersect(b)
filter
BedTool.filter() 可以对BedTool 对象进行过滤。传递一个函数,其接收的第一个参数是一个Interval. 返回True/False来进行过滤。
a = pybedtools.example_bedtool('a.bed')
b = a.filter(lambda x: len(x) > 100)
print(b)
# chr1 150 500 feature3 0 -
bed= BedTool(bedfile)
filtered = bed.filter(lambda a: a.name in snp_list)bed = bed.filter(lambda r: r.chrom == chrom)
iltered_ref = full_ref.filter(lambda gtf: gtf[2] == args.feature)self.bedtool.filter(lambda x : x[0] == 'chr' + str(int(chrnum))).saveas()
sort
第一列升序排序(按照字符;染色体号),第二列按照数字排序(起始位置);
bedtools merge requires that you presort your data by chromosome and then by start position (e.g., sort -k1,1 -k2,2n in.bed > in.sorted.bed for BED files).
这个命令这样写也许看的更清楚些: sort -k 1,1 -k 2,2n input.bed > input_sorted.bed
主要有两个参数 -n 和 -k;
其中 -n 的意思是按照数值大小排序。(-n 参数在 2,2 后面, 它仅对第二列有效)
其中 -k 的意思是指定排序的列(域),比如说 -k 1,1 表示仅仅对第一列有效, -k 2,2n 表示仅仅对第二列按照数字排序
其中 -t 的意思是指定行分隔符, 这里应该是 -t $'\t', 因为默认的也是, 所以可以省略。
cat
Concatenate interval files together.
Concatenates two BedTool objects (or an object and a file) and does an optional post-merge of the features.
postmerge=True by default; use postmerge=False if you want to keep features separate.
force_truncate=False by default; force_truncate=True to truncate all files to chrom, start, stop.
When force_truncate=False and postmerge=False, the output will contain the smallest number of fields observed across all inputs. This maintains compatibility with BEDTools programs, which assume constant number of fields in all lines of a file.
Other kwargs are sent to BedTool.merge() (and assuming that postmerge=True).
a = pybedtools.example_bedtool('a.bed')
b = pybedtools.example_bedtool('b.bed')
print(a.cat(b))
chr1 1 500
chr1 800 950
print(a.cat(*[b,b],
postmerge=False))
chr1 1 100 feature1 0 +
chr1 100 200 feature2 0 +
chr1 150 500 feature3 0 -
chr1 900 950 feature4 0 +
chr1 155 200 feature5 0 -
chr1 800 901 feature6 0 +
chr1 155 200 feature5 0 -
chr1 800 901 feature6 0 +
REF: https://daler.github.io/pybedtools/autodocs/pybedtools.bedtool.BedTool.cat.html
each
BedTool.each()也是将函数应用于每个Interval, 但主要是对Interval进行修改。
slop
Add requested base pairs of "slop" to each feature.
REF: https://bedtools.readthedocs.io/en/latest/content/tools/slop.html


Usage and option summary
Usage:
bedtools slop [OPTIONS] -i <BED/GFF/VCF> -g <GENOME> [-b or (-l and -r)]
(or):
slopBed [OPTIONS] -i <BED/GFF/VCF> -g <GENOME> [-b or (-l and -r)]
| Option | Description |
|---|---|
| -b | Increase the BED/GFF/VCF entry by the same number base pairs in each direction. Integer. |
| -l | The number of base pairs to subtract from the start coordinate. Integer. |
| -r | The number of base pairs to add to the end coordinate. Integer. |
| -s | Define -l and -r based on strand. For example. if used, -l 500 for a negative-stranded feature, it will add 500 bp to the end coordinate. |
| -pct | Define -l and -r as a fraction of the feature’s length. E.g. if used on a 1000bp feature, -l 0.50, will add 500 bp “upstream”. Default = false. |
| -header | Print the header from the input file prior to results. |
Default behavior
By default, bedtools slop will either add a fixed number of bases in each direction (-b) or an asymmetric number of bases in each direction with -l and -r.
$ cat A.bed chr1 5 100 chr1 800 980 $ cat my.genome chr1 1000 $ bedtools slop -i A.bed -g my.genome -b 5 chr1 0 105 chr1 795 985 $ bedtools slop -i A.bed -g my.genome -l 2 -r 3 chr1 3 103 chr1 798 983
However, if the requested number of bases exceeds the boundaries of the chromosome, bedtools slop will “clip” the feature accordingly.
$ cat A.bed chr1 5 100 chr1 800 980 $ cat my.genome chr1 1000 $ bedtools slop -i A.bed -g my.genome -b 5000 chr1 0 1000 chr1 0 1000
-s Resizing features according to strand
bedtools slop will optionally increase the size of a feature based on strand.
For example:
$ cat A.bed chr1 100 200 a1 1 + chr1 100 200 a2 2 - $ cat my.genome chr1 1000 $ bedtools slop -i A.bed -g my.genome -l 50 -r 80 -s chr1 50 280 a1 1 + chr1 20 250 a2 2 -
-pct Resizing features by a given fraction
bedtools slop will optionally increase the size of a feature by a user-specific fraction.
For example:
$ cat A.bed chr1 100 200 a1 1 + $ bedtools slop -i A.bed -g my.genome -b 0.5 -pct chr1 50 250 a1 1 + $ bedtools slop -i a.bed -l 0.5 -r 0.0 -pct -g my.genome chr1 50 200 a1 1 +
-header Print the header for the A file before reporting results.
By default, if your A file has a header, it is ignored when reporting results. This option will instead tell bedtools to first print the header for the A file prior to reporting results.
window_maker
There are two alternatives for supplying a genome. Use g="genome.filename" if you have a genome’s chrom sizes saved as a file. This is the what BEDTools expects when using it from the command line. Alternatively, use the genome="assembly.name" (for example, genome="hg19") to use chrom sizes for that assembly without having to manage a separate file. The genome argument triggers a call pybedtools.chromsizes, so see that method for more details.
REF https://daler.github.io/pybedtools/autodocs/pybedtools.bedtool.BedTool.window_maker.html
Tool: bedtools makewindows
Version: v2.30.0
Summary: Makes adjacent or sliding windows across a genome or BED file.
Usage: bedtools makewindows [OPTIONS] [-g <genome> OR -b <bed>]
[ -w <window**size> OR -n <number of windows> ]
Input Options:
-g <genome>
Genome file size (see notes below).
Windows will be created for each chromosome in the file.
-b <bed>
BED file (with chrom,start,end fields).
Windows will be created for each interval in the file.
Windows Output Options:
-w <window**size>
Divide each input interval (either a chromosome or a BED interval)
to fixed-sized windows (i.e. same number of nucleotide in each window).
Can be combined with -s <step**size>
-s <step**size>
Step size: i.e., how many base pairs to step before
creating a new window. Used to create "sliding" windows.
- Defaults to window size (non-sliding windows).
-n <number**of**windows>
Divide each input interval (either a chromosome or a BED interval)
to fixed number of windows (i.e. same number of windows, with
varying window sizes).
-reverse
Reverse numbering of windows in the output, i.e. report
windows in decreasing order
ID Naming Options:
-i src|winnum|srcwinnum
The default output is 3 columns: chrom, start, end .
With this option, a name column will be added.
"-i src" - use the source interval's name.
"-i winnum" - use the window number as the ID (e.g. 1,2,3,4...).
"-i srcwinnum" - use the source interval's name with the window number.
See below for usage examples.
Notes:
(1) The genome file should tab delimited and structured as follows:
<chromName><TAB><chromSize>
For example, Human (hg19):
chr1 249250621
chr2 243199373
...
chr18**gl000207**random 4262
Tips:
One can use the UCSC Genome Browser's MySQL database to extract
chromosome sizes. For example, H. sapiens:
mysql --user=genome --host=genome-mysql.cse.ucsc.edu -A -e \
"select chrom, size from hg19.chromInfo" > hg19.genome
Examples:
# Divide the human genome into windows of 1MB:
$ bedtools makewindows -g hg19.txt -w 1000000
chr1 0 1000000
chr1 1000000 2000000
chr1 2000000 3000000
chr1 3000000 4000000
chr1 4000000 5000000
...
# Divide the human genome into sliding (=overlapping) windows of 1MB, with 500KB overlap:
$ bedtools makewindows -g hg19.txt -w 1000000 -s 500000
chr1 0 1000000
chr1 500000 1500000
chr1 1000000 2000000
chr1 1500000 2500000
chr1 2000000 3000000
...
# Divide each chromosome in human genome to 1000 windows of equal size:
$ bedtools makewindows -g hg19.txt -n 1000
chr1 0 249251
chr1 249251 498502
chr1 498502 747753
chr1 747753 997004
chr1 997004 1246255
...
# Divide each interval in the given BED file into 10 equal-sized windows:
$ cat input.bed
chr5 60000 70000
chr5 73000 90000
chr5 100000 101000
$ bedtools makewindows -b input.bed -n 10
chr5 60000 61000
chr5 61000 62000
chr5 62000 63000
chr5 63000 64000
chr5 64000 65000
...
# Add a name column, based on the window number:
$ cat input.bed
chr5 60000 70000 AAA
chr5 73000 90000 BBB
chr5 100000 101000 CCC
$ bedtools makewindows -b input.bed -n 3 -i winnum
chr5 60000 63334 1
chr5 63334 66668 2
chr5 66668 70000 3
chr5 73000 78667 1
chr5 78667 84334 2
chr5 84334 90000 3
chr5 100000 100334 1
chr5 100334 100668 2
chr5 100668 101000 3
...
# Reverse window numbers:
$ cat input.bed
chr5 60000 70000 AAA
chr5 73000 90000 BBB
chr5 100000 101000 CCC
$ bedtools makewindows -b input.bed -n 3 -i winnum -reverse
chr5 60000 63334 3
chr5 63334 66668 2
chr5 66668 70000 1
chr5 73000 78667 3
chr5 78667 84334 2
chr5 84334 90000 1
chr5 100000 100334 3
chr5 100334 100668 2
chr5 100668 101000 1
...
# Add a name column, based on the source ID + window number:
$ cat input.bed
chr5 60000 70000 AAA
chr5 73000 90000 BBB
chr5 100000 101000 CCC
$ bedtools makewindows -b input.bed -n 3 -i srcwinnum
chr5 60000 63334 AAA**1
chr5 63334 66668 AAA**2
chr5 66668 70000 AAA**3
chr5 73000 78667 BBB**1
chr5 78667 84334 BBB**2
chr5 84334 90000 BBB**3
chr5 100000 100334 CCC**1
chr5 100334 100668 CCC**2
chr5 100668 101000 CCC**3
...
merge() 合并坐标区间
x1 = a.intersect(b, u=True)
x2 = x1.merge()注意:合并之前必须先排序sort。
将一个BED文件中所有的重叠区域和相邻merge为一个大的区域,可以将结果转换为其他格式的数据,可以通过参数设置相距多远的两个区域,可以取他们的并集:

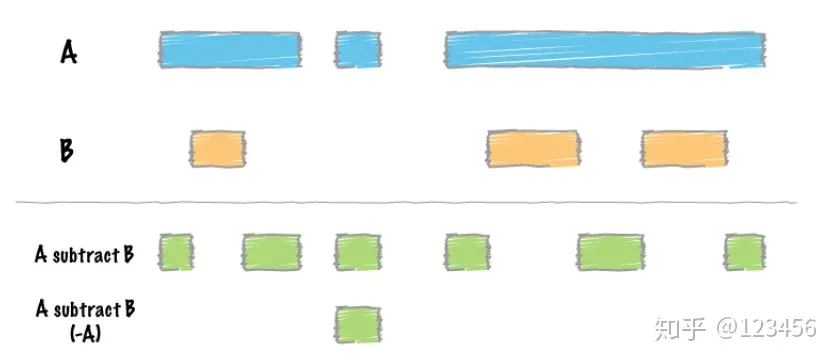
差集运算,用A中的区域减去B中的区域,通过参数-A可以实现有重叠区域的全部剔除:
Interval
在pybedtools中, 以Interval对象来表示BED,GFF,GTF或VCF文件中的一行数据。
REF
https://zhuanlan.zhihu.com/p/52322803
REF
https://www.jianshu.com/p/183fe949aca7
标签:chr5,chr1,pybedtools,bed,genome,使用,bedtools,安装 From: https://www.cnblogs.com/emanlee/p/17073574.html